RNA interference (RNAi) screening approach identifies agents that enhance paclitaxel activity in breast cancer cells
- PMID: 20576088
- PMCID: PMC2917036
- DOI: 10.1186/bcr2595
RNA interference (RNAi) screening approach identifies agents that enhance paclitaxel activity in breast cancer cells
Abstract
Introduction: Paclitaxel is a widely used drug in the treatment of patients with locally advanced and metastatic breast cancer. However, only a small portion of patients have a complete response to paclitaxel-based chemotherapy, and many patients are resistant. Strategies that increase sensitivity and limit resistance to paclitaxel would be of clinical use, especially for patients with triple-negative breast cancer (TNBC).
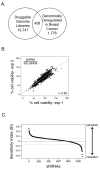
Methods: We generated a gene set from overlay of the druggable genome and a collection of genomically deregulated gene transcripts in breast cancer. We used loss-of-function RNA interference (RNAi) to identify gene products in this set that, when targeted, increase paclitaxel sensitivity. Pharmacological agents that targeted the top scoring hits/genes from our RNAi screens were used in combination with paclitaxel, and the effects on the growth of various breast cancer cell lines were determined.
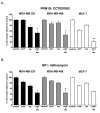
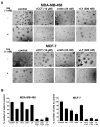
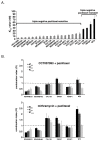
Results: RNAi screens performed herein were validated by identification of genes in pathways that, when previously targeted, enhanced paclitaxel sensitivity in the pre-clinical and clinical settings. When chemical inhibitors, CCT007093 and mithramycin, against two top hits in our screen, PPMID and SP1, respectively, were used in combination with paclitaxel, we observed synergistic growth inhibition in both 2D and 3D breast cancer cell cultures. The transforming growth factor beta (TGFbeta) receptor inhibitor, LY2109761, that targets the signaling pathway of another top scoring hit, TGFbeta1, was synergistic with paclitaxel when used in combination on select breast cancer cell lines grown in 3D culture. We also determined the relative paclitaxel sensitivity of 22 TNBC cell lines and identified 18 drug-sensitive and four drug-resistant cell lines. Of significance, we found that both CCT007093 and mithramycin, when used in combination with paclitaxel, resulted in synergistic inhibition of the four paclitaxel-resistant TNBC cell lines.
Conclusions: RNAi screening can identify druggable targets and novel drug combinations that can sensitize breast cancer cells to paclitaxel. This genomic-based approach can be applied to a multitude of tumor-derived cell lines and drug treatments to generate requisite pre-clinical data for new drug combination therapies to pursue in clinical investigations.
Figures
Similar articles
-
An in vivo genome-wide shRNA screen identifies BCL6 as a targetable biomarker of paclitaxel resistance in breast cancer.Mol Oncol. 2021 Aug;15(8):2046-2064. doi: 10.1002/1878-0261.12964. Epub 2021 May 18. Mol Oncol. 2021. PMID: 33932086 Free PMC article.
-
Analysis of high-throughput RNAi screening data in identifying genes mediating sensitivity to chemotherapeutic drugs: statistical approaches and perspectives.BMC Genomics. 2012;13 Suppl 8(Suppl 8):S3. doi: 10.1186/1471-2164-13-S8-S3. Epub 2012 Dec 17. BMC Genomics. 2012. PMID: 23281588 Free PMC article.
-
Pharmacological and small interference RNA-mediated inhibition of breast cancer-associated fatty acid synthase (oncogenic antigen-519) synergistically enhances Taxol (paclitaxel)-induced cytotoxicity.Int J Cancer. 2005 May 20;115(1):19-35. doi: 10.1002/ijc.20754. Int J Cancer. 2005. PMID: 15657900
-
Paclitaxel combination therapy in the treatment of metastatic breast cancer: a review.Semin Oncol. 1996 Oct;23(5 Suppl 11):46-56. Semin Oncol. 1996. PMID: 8893900 Review.
-
Paclitaxel resistance in breast cancer: Current challenges and recent advanced therapeutic strategies.Cancer Treat Res Commun. 2025;43:100918. doi: 10.1016/j.ctarc.2025.100918. Epub 2025 Mar 31. Cancer Treat Res Commun. 2025. PMID: 40215760 Review.
Cited by
-
Prognostic risk assessment model and drug sensitivity analysis of colon adenocarcinoma (COAD) based on immune-related lncRNA pairs.BMC Bioinformatics. 2022 Oct 18;23(1):435. doi: 10.1186/s12859-022-04969-4. BMC Bioinformatics. 2022. PMID: 36258178 Free PMC article.
-
Three-dimensional in vitro co-culture model of breast tumor using magnetic levitation.Sci Rep. 2014 Oct 1;4:6468. doi: 10.1038/srep06468. Sci Rep. 2014. PMID: 25270048 Free PMC article.
-
Identifying Therapies to Combat Epithelial Mesenchymal Plasticity-Associated Chemoresistance to Conventional Breast Cancer Therapies Using An shRNA Library Screen.Cancers (Basel). 2020 Apr 30;12(5):1123. doi: 10.3390/cancers12051123. Cancers (Basel). 2020. PMID: 32365878 Free PMC article.
-
Targeting of apoptotic pathways by SMAC or BH3 mimetics distinctly sensitizes paclitaxel-resistant triple negative breast cancer cells.Oncotarget. 2017 Jul 11;8(28):45088-45104. doi: 10.18632/oncotarget.15125. Oncotarget. 2017. PMID: 28187446 Free PMC article.
-
Fe-doped chrysotile nanotubes containing siRNAs to silence SPAG5 to treat bladder cancer.J Nanobiotechnology. 2021 Jun 23;19(1):189. doi: 10.1186/s12951-021-00935-z. J Nanobiotechnology. 2021. PMID: 34162370 Free PMC article.
References
-
- Hortobagyi G. Docetaxel in breast cancer and a rationale for combination therapy. Oncology (Williston Park) 1997;11:11–15. - PubMed
-
- Hortobagyi GN. Paclitaxel-based combination chemotherapy for breast cancer. Oncology (Williston Park) 1997;11:29–37. - PubMed
-
- Ayers M, Symmans WF, Stec J, Damokosh AI, Clark E, Hess K, Lecocke M, Metivier J, Booser D, Ibrahim N, Valero V, Royce M, Arun B, Whitman G, Ross J, Sneige N, Hortobagyi GN, Pusztai L. Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. J Clin Oncol. 2004;22:2284–2293. doi: 10.1200/JCO.2004.05.166. - DOI - PubMed
-
- Dressman HK, Hans C, Bild A, Olson JA, Rosen E, Marcom PK, Liotcheva VB, Jones EL, Vujaskovic Z, Marks J, Dewhirst MW, West M, Nevins JR, Blackwell K. Gene expression profiles of multiple breast cancer phenotypes and response to neoadjuvant chemotherapy. Clin Cancer Res. 2006;12:819–826. doi: 10.1158/1078-0432.CCR-05-1447. - DOI - PubMed
-
- Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ, Rouzier R, Sneige N, Ross JS, Vidaurre T, Gomez HL, Hortobagyi GN, Pusztai L. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

