Alternative ways to think about cellular internal ribosome entry
- PMID: 20576611
- PMCID: PMC2937932
- DOI: 10.1074/jbc.R110.150532
Alternative ways to think about cellular internal ribosome entry
Abstract
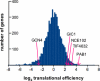
Internal ribosome entry sites (IRESs) are specialized mRNA elements that allow recruitment of eukaryotic ribosomes to naturally uncapped mRNAs or to capped mRNAs under conditions in which cap-dependent translation is inhibited. Putative cellular IRESs have been proposed to play crucial roles in stress responses, development, apoptosis, cell cycle control, and neuronal function. However, most of the evidence for cellular IRES activity rests on bicistronic reporter assays, the reliability of which has been questioned. Here, the mechanisms underlying cap-independent translation of cellular mRNAs and the contributions of such translation to cellular protein synthesis are discussed. I suggest that the division of cellular mRNAs into mutually exclusive categories of "cap-dependent" and "IRES-dependent" should be reconsidered and that the implications of cellular IRES activity need to be incorporated into our models of cap-dependent initiation.
Figures

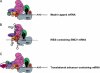
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

