Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales
- PMID: 20576738
- PMCID: PMC2924827
- DOI: 10.1093/aob/mcq129
Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales
Abstract
Background and aims: To assess the number and phylogenetic distribution of large-scale genome duplications in the ancestry of Actinidia, publicly available expressed sequenced tags (ESTs) for members of the Actinidiaceae and related Ericales, including tea (Camellia sinensis), were analysed.
Methods: Synonymous divergences (K(s)) were calculated for all duplications within gene families and examined for evidence of large-scale duplication events. Phylogenetic comparisons for a selection of orthologues among several related species in Ericales and two outgroups permitted placement of duplication events in relation to lineage divergences. Gene ontology (GO) categories were analysed for each whole-genome duplication (WGD) and the whole transcriptome.
Key results: Evidence for three ancient WGDs in Actinidia was found. Analyses of paleologue GO categories indicated a different pattern of retained genes for each genome duplication, but a pattern consistent with the dosage-balance hypothesis among all retained paleologues.
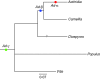
Conclusions: This study provides evidence for one independent WGD in the ancestry of Actinidia (Ad-alpha), a WGD shared by Actinidia and Camellia (Ad-beta), and the well-established At-gamma WGD that occurred prior to the divergence of all taxa examined. More ESTs in other taxa are needed to elucidate which groups in Ericales share the Ad-beta or Ad-alpha duplications and their impact on diversification.
Figures


Similar articles
-
Draft genome of the kiwifruit Actinidia chinensis.Nat Commun. 2013;4:2640. doi: 10.1038/ncomms3640. Nat Commun. 2013. PMID: 24136039 Free PMC article.
-
Phylotranscriptomics of Theaceae: generic-level relationships, reticulation and whole-genome duplication.Ann Bot. 2022 Mar 23;129(4):457-471. doi: 10.1093/aob/mcac007. Ann Bot. 2022. PMID: 35037017 Free PMC article.
-
The First Complete Chloroplast Genome Sequences in Actinidiaceae: Genome Structure and Comparative Analysis.PLoS One. 2015 Jun 5;10(6):e0129347. doi: 10.1371/journal.pone.0129347. eCollection 2015. PLoS One. 2015. PMID: 26046631 Free PMC article.
-
Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model.Curr Opin Plant Biol. 2012 Apr;15(2):147-53. doi: 10.1016/j.pbi.2012.03.011. Epub 2012 Apr 3. Curr Opin Plant Biol. 2012. PMID: 22480429 Review.
-
Functional Divergence between Subgenomes and Gene Pairs after Whole Genome Duplications.Mol Plant. 2018 Mar 5;11(3):388-397. doi: 10.1016/j.molp.2017.12.010. Epub 2017 Dec 22. Mol Plant. 2018. PMID: 29275166 Review.
Cited by
-
The Rhododendron Genome and Chromosomal Organization Provide Insight into Shared Whole-Genome Duplications across the Heath Family (Ericaceae).Genome Biol Evol. 2019 Dec 1;11(12):3353-3371. doi: 10.1093/gbe/evz245. Genome Biol Evol. 2019. PMID: 31702783 Free PMC article.
-
Evidence of multiple genome duplication events in Mytilus evolution.BMC Genomics. 2022 May 2;23(1):340. doi: 10.1186/s12864-022-08575-9. BMC Genomics. 2022. PMID: 35501689 Free PMC article.
-
Genes in evolution: the control of diversity and speciation.Ann Bot. 2010 Sep;106(3):437-8. doi: 10.1093/aob/mcq168. Ann Bot. 2010. PMID: 20729371 Free PMC article. No abstract available.
-
GOgetter: A pipeline for summarizing and visualizing GO slim annotations for plant genetic data.Appl Plant Sci. 2023 Aug 11;11(4):e11536. doi: 10.1002/aps3.11536. eCollection 2023 Jul-Aug. Appl Plant Sci. 2023. PMID: 37601315 Free PMC article.
-
Phylogenetic pinpointing of a paleopolyploidy event within the flax genus (Linum) using transcriptomics.Ann Bot. 2014 Apr;113(5):753-61. doi: 10.1093/aob/mct306. Epub 2013 Dec 30. Ann Bot. 2014. PMID: 24380843 Free PMC article.
References
-
- Anderberg AA, Rydin C, Källersjö M. Phylogenetic relationships in the order Ericales s.l.: analyses of molecular data from five genes from the plastid and mitochondrial genomes. American Journal of Botany. 2002;89:677–687. - PubMed
-
- Barker MS, Wolf PG. Unfurling fern biology in the genomics age. BioScience. 2010;60:177–185.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

