Large Deformation Diffeomorphic Metric Mapping Registration of Reconstructed 3D Histological Section Images and in vivo MR Images
- PMID: 20577633
- PMCID: PMC2889720
- DOI: 10.3389/fnhum.2010.00043
Large Deformation Diffeomorphic Metric Mapping Registration of Reconstructed 3D Histological Section Images and in vivo MR Images
Abstract
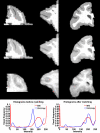
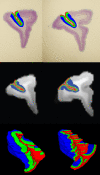
Our current understanding of neuroanatomical abnormalities in neuropsychiatric diseases is based largely on magnetic resonance imaging (MRI) and post mortem histological analyses of the brain. Further advances in elucidating altered brain structure in these human conditions might emerge from combining MRI and histological methods. We propose a multistage method for registering 3D volumes reconstructed from histological sections to corresponding in vivo MRI volumes from the same subjects: (1) manual segmentation of white matter (WM), gray matter (GM) and cerebrospinal fluid (CSF) compartments in histological sections, (2) alignment of consecutive histological sections using 2D rigid transformation to construct a 3D histological image volume from the aligned sections, (3) registration of reconstructed 3D histological volumes to the corresponding 3D MRI volumes using 3D affine transformation, (4) intensity normalization of images via histogram matching, and (5) registration of the volumes via intensity based large deformation diffeomorphic metric (LDDMM) image matching algorithm. Here we demonstrate the utility of our method in the transfer of cytoarchitectonic information from histological sections to identify regions of interest in MRI scans of nine adult macaque brains for morphometric analyses. LDDMM improved the accuracy of the registration via decreased distances between GM/CSF surfaces after LDDMM (0.39 +/- 0.13 mm) compared to distances after affine registration (0.76 +/- 0.41 mm). Similarly, WM/GM distances decreased to 0.28 +/- 0.16 mm after LDDMM compared to 0.54 +/- 0.39 mm after affine registration. The multistage registration method may find broad application for mapping histologically based information, for example, receptor distributions, gene expression, onto MRI volumes.
Keywords: LDDMM; MRI; affine registration; area 46; histology; nonlinear registration; prefrontal cortex.
Figures


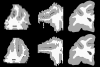

References
-
- Andreasen A., Drewes A. M., Assentoft J. E., Larsen N. E. (1992). Computer-assisted alignment of standard serial sections without use of artificial landmarks. A practical approach to the utilization of incomplete information in 3-D reconstruction of the hippocampal region. J. Neurosci. Methods 45, 199–20710.1016/0165-0270(92)90077-Q - DOI - PubMed
-
- Bardinet E., Ourselin S., Dormont D., Malandain G., Tandé D., Parain K., Ayache N., Yelnik J. (2002). Co-registration of histological, optical and MR data of the human brain. LNCS 2488, 548–555
-
- Beg M. F., Miller M. I., Trouvé A., Younes L. (2005). Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int. J. Comput. Vision 61, 139–15710.1023/B:VISI.0000043755.93987.aa - DOI
-
- Boothby W. M. (2002). An Introduction to Differentiable Manifolds and Riemannian Geometry, 2nd edn.New York: Academic Press
Grants and funding
LinkOut - more resources
Full Text Sources

