Hepatocyte NAD(P)H oxidases as an endogenous source of reactive oxygen species during hepatitis C virus infection
- PMID: 20578128
- PMCID: PMC3141587
- DOI: 10.1002/hep.23671
Hepatocyte NAD(P)H oxidases as an endogenous source of reactive oxygen species during hepatitis C virus infection
Abstract
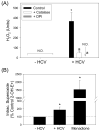
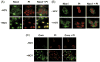
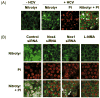
Oxidative stress has been identified as a key mechanism of hepatitis C virus (HCV)-induced pathogenesis. Studies have suggested that HCV increases the generation of hydroxyl radical and peroxynitrite close to the cell nucleus, inflicting DNA damage, but the source of reactive oxygen species (ROS) remains incompletely characterized. We hypothesized that HCV increases the generation of superoxide and hydrogen peroxide close to the hepatocyte nucleus and that this source of ROS is reduced nicotinamide adenine dinucleotide phosphate (NAD(P)H) oxidase 4 (Nox4). Huh7 human hepatoma cells and telomerase-reconstituted primary human hepatocytes, transfected or infected with virus-producing HCV strains of genotypes 2a and 1b, were examined for messenger RNA (mRNA), protein, and subcellular localization of Nox proteins along with the human liver. We found that genotype 2a HCV induced persistent elevations of Nox1 and Nox4 mRNA and proteins in Huh7 cells. HCV genotype 1b likewise elevated the levels of Nox1 and Nox4 in telomerase-reconstituted primary human hepatocytes. Furthermore, Nox1 and Nox4 proteins were increased in HCV-infected human liver versus uninfected liver samples. Unlike Nox1, Nox4 was prominent in the nuclear compartment of these cells as well as the human liver, particularly in the presence of HCV. HCV-induced ROS and nuclear nitrotyrosine could be decreased with small interfering RNAs to Nox1 and Nox4. Finally, HCV increased the level of transforming growth factor beta 1 (TGFbeta1). TGFbeta1 could elevate Nox4 expression in the presence of infectious HCV, and HCV increased Nox4 at least in part through TGFbeta1.
Conclusion: HCV induced a persistent elevation of Nox1 and Nox4 and increased nuclear localization of Nox4 in hepatocytes in vitro and in the human liver. Hepatocyte Nox proteins are likely to act as a persistent, endogenous source of ROS during HCV-induced pathogenesis.
Figures











References
-
- Seronello S, Sheikh MY, Choi J. Redox regulation of hepatitis C in nonalcoholic and alcoholic liver. Free Radic Biol Med. 2007;43:869–882. - PubMed
-
- Garcia-Monzon C, Majano PL, Zubia I, Sanz P, Apolinario A, Moreno-Otero R. Intrahepatic accumulation of nitrotyrosine in chronic viral hepatitis is associated with histological severity of liver disease. J Hepatol. 2000;32:331–338. - PubMed
-
- Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. - PubMed
-
- Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J Physiol. 1996;271:C1424–1437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
