Population genetic structure of the people of Qatar
- PMID: 20579625
- PMCID: PMC2896773
- DOI: 10.1016/j.ajhg.2010.05.018
Population genetic structure of the people of Qatar
Abstract
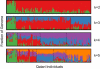
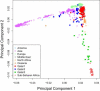
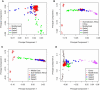
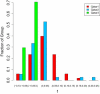
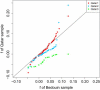
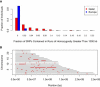
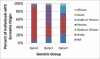
People of the Qatar peninsula represent a relatively recent founding by a small number of families from three tribes of the Arabian Peninsula, Persia, and Oman, with indications of African admixture. To assess the roles of both this founding effect and the customary first-cousin marriages among the ancestral Islamic populations in Qatar's population genetic structure, we obtained and genotyped with Affymetrix 500k SNP arrays DNA samples from 168 self-reported Qatari nationals sampled from Doha, Qatar. Principal components analysis was performed along with samples from the Human Genetic Diversity Project data set, revealing three clear clusters of genotypes whose proximity to other human population samples is consistent with Arabian origin, a more eastern or Persian origin, and individuals with African admixture. The extent of linkage disequilibrium (LD) is greater than that of African populations, and runs of homozygosity in some individuals reflect substantial consanguinity. However, the variance in runs of homozygosity is exceptionally high, and the degree of identity-by-descent sharing generally appears to be lower than expected for a population in which nearly half of marriages are between first cousins. Despite the fact that the SNPs of the Affymetrix 500k chip were ascertained with a bias toward SNPs common in Europeans, the data strongly support the notion that the Qatari population could provide a valuable resource for the mapping of genes associated with complex disorders and that tests of pairwise interactions are particularly empowered by populations with elevated LD like the Qatari.
Copyright 2010 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G., Wellcome Trust Case Control Consortium Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. - PMC - PubMed
-
- Meyre D., Delplanque J., Chèvre J.C., Lecoeur C., Lobbens S., Gallina S., Durand E., Vatin V., Degraeve F., Proença C. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 2009;41:157–159. - PubMed
-
- Badii R., Bener A., Zirie M., Al-Rikabi A., Simsek M., Al-Hamaq A.O., Ghoussaini M., Froguel P., Wareham N.J. Lack of association between the Pro12Ala polymorphism of the PPAR-gamma 2 gene and type 2 diabetes mellitus in the Qatari consanguineous population. Acta Diabetol. 2008;45:15–21. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

