Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection
- PMID: 20580051
- PMCID: PMC2925245
- DOI: 10.1016/j.virol.2010.05.032
Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection
Abstract
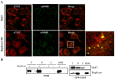
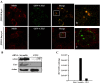
Autophagy is an important cellular process by which ATG5 initiates the formation of double membrane vesicles (DMVs). Upon infection, DMVs have been shown to harbor the replicase complex of positive-strand RNA viruses such as MHV, poliovirus, and equine arteritis virus. Recently, it has been shown that autophagy proteins are proviral factors that favor initiation of hepatitis C virus (HCV) infection. Here, we identified ATG5 as an interacting protein for the HCV NS5B. ATG5/NS5B interaction was confirmed by co-IP and metabolic labeling studies. Furthermore, ATG5 protein colocalizes with NS4B, a constituent of the membranous web. Importantly, immunofluorescence staining demonstrated a strong colocalization of ATG5 and NS5B within perinuclear regions of infected cells at 2 days postinfection. However, colocalization was completely lacking at 5DPI, suggesting that HCV utilizes ATG5 as a proviral factor during the onset of viral infection. Finally, inhibition of autophagy through ATG5 silencing blocks HCV replication.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
The autophagy elongation complex (ATG5-12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation.Sci Rep. 2017 Jan 9;7:40351. doi: 10.1038/srep40351. Sci Rep. 2017. PMID: 28067309 Free PMC article.
-
LC3B is not recruited along with the autophagy elongation complex (ATG5-12/16L1) at HCV replication site and is dispensable for viral replication.PLoS One. 2018 Oct 4;13(10):e0205189. doi: 10.1371/journal.pone.0205189. eCollection 2018. PLoS One. 2018. PMID: 30286180 Free PMC article.
-
Surfeit 4 Contributes to the Replication of Hepatitis C Virus Using Double-Membrane Vesicles.J Virol. 2020 Jan 6;94(2):e00858-19. doi: 10.1128/JVI.00858-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31645450 Free PMC article.
-
Structures of hepatitis C virus nonstructural proteins required for replicase assembly and function.Curr Opin Virol. 2013 Apr;3(2):129-36. doi: 10.1016/j.coviro.2013.03.013. Epub 2013 Apr 16. Curr Opin Virol. 2013. PMID: 23601958 Free PMC article. Review.
-
Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase).Curr Top Microbiol Immunol. 2000;242:225-60. doi: 10.1007/978-3-642-59605-6_11. Curr Top Microbiol Immunol. 2000. PMID: 10592663 Review. No abstract available.
Cited by
-
Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication.PLoS Pathog. 2012;8(12):e1003056. doi: 10.1371/journal.ppat.1003056. Epub 2012 Dec 6. PLoS Pathog. 2012. PMID: 23236278 Free PMC article.
-
Mitophagy in the Pathogenesis of Liver Diseases.Cells. 2020 Mar 30;9(4):831. doi: 10.3390/cells9040831. Cells. 2020. PMID: 32235615 Free PMC article. Review.
-
HCV and Oxidative Stress: Implications for HCV Life Cycle and HCV-Associated Pathogenesis.Oxid Med Cell Longev. 2016;2016:9012580. doi: 10.1155/2016/9012580. Epub 2016 Feb 3. Oxid Med Cell Longev. 2016. PMID: 26955431 Free PMC article. Review.
-
The autophagy elongation complex (ATG5-12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation.Sci Rep. 2017 Jan 9;7:40351. doi: 10.1038/srep40351. Sci Rep. 2017. PMID: 28067309 Free PMC article.
-
Cellular factors involved in the hepatitis C virus life cycle.World J Gastroenterol. 2021 Jul 28;27(28):4555-4581. doi: 10.3748/wjg.v27.i28.4555. World J Gastroenterol. 2021. PMID: 34366623 Free PMC article. Review.
References
-
- Aizaki H., Choi K.S., Liu M., Li Y.J., Lai M.M. Polypyrimidine-tract-binding protein is a component of the HCV RNA replication complex and necessary for RNA synthesis. J. Biomed. Sci. 2006;13(4):469–480. - PubMed
-
- Dreux M., Chisari F.V. Autophagy proteins promote hepatitis C virus replication. Autophagy. 2009;5(8):1224–1225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

