Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein
- PMID: 20580052
- PMCID: PMC2914208
- DOI: 10.1016/j.virol.2010.05.031
Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein
Abstract
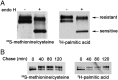
Coronaviruses are enveloped RNA viruses that generally cause mild disease in humans. However, the recently emerged coronavirus that caused severe acute respiratory syndrome (SARS-CoV) is the most pathogenic human coronavirus discovered to date. The SARS-CoV spike (S) protein mediates virus entry by binding cellular receptors and inducing fusion between the viral envelope and the host cell membrane. Coronavirus S proteins are palmitoylated, which may affect function. Here, we created a non-palmitoylated SARS-CoV S protein by mutating all nine cytoplasmic cysteine residues. Palmitoylation of SARS-CoV S was required for partitioning into detergent-resistant membranes and for cell-cell fusion. Surprisingly, however, palmitoylation of S was not required for interaction with SARS-CoV M protein. This contrasts with the requirement for palmitoylation of mouse hepatitis virus S protein for interaction with M protein and may point to important differences in assembly and infectivity of these two coronaviruses.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures



References
-
- Baker T.L., Zheng H., Walker J., Coloff J.L., Buss J.E. Distinct rates of palmitate turnover on membrane-bound cellular and oncogenic H-ras. J. Biol. Chem. 2003;278(21):19292–19300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

