Comparison of multivariate classifiers and response normalizations for pattern-information fMRI
- PMID: 20580933
- PMCID: PMC2914143
- DOI: 10.1016/j.neuroimage.2010.05.051
Comparison of multivariate classifiers and response normalizations for pattern-information fMRI
Abstract
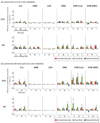
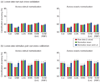
A popular method for investigating whether stimulus information is present in fMRI response patterns is to attempt to "decode" the stimuli from the response patterns with a multivariate classifier. The sensitivity for detecting the information depends on the particular classifier used. However, little is known about the relative performance of different classifiers on fMRI data. Here we compared six multivariate classifiers and investigated how the response-amplitude estimate used (beta- or t-value) and different pattern normalizations affect classification performance. The compared classifiers were a pattern-correlation classifier, a k-nearest-neighbors classifier, Fisher's linear discriminant, Gaussian naïve Bayes, and linear and nonlinear (radial-basis-function kernel) support vector machines. We compared these classifiers' accuracy at decoding the category of visual objects from response patterns in human early visual and inferior temporal cortex acquired in an event-related design with BOLD fMRI at 3T using SENSE and isotropic voxels of about 2-mm width. Overall, Fisher's linear discriminant (with an optimal-shrinkage covariance estimator) and the linear support vector machine performed best. The pattern-correlation classifier often performed similarly as those two classifiers. The nonlinear classifiers never performed better and sometimes significantly worse than the linear classifiers, suggesting overfitting. Defining response patterns by t-values (or in error-standard-deviation units) rather than by beta estimates (in % signal change) to define the patterns appeared advantageous. Cross-validation by a leave-one-stimulus-pair-out method gave higher accuracies than a leave-one-run-out method, suggesting that generalization to independent runs (which more safely ensures independence of the test set) is more challenging than generalization to novel stimuli within the same category. Independent selection of fewer more visually responsive voxels tended to yield better decoding performance for all classifiers. Normalizing mean and standard deviation of the response patterns either across stimuli or across voxels had no significant effect on decoding performance. Overall our results suggest that linear decoders based on t-value patterns may perform best in the present scenario of visual object representations measured for about 60min per subject with 3T fMRI.
Published by Elsevier Inc.
Figures





References
-
- Bishop CM. Pattern Recognition and Machine Learning. New York: Springer; 2007.
-
- Björnsdotter Åberg M, Löken L, Wessberg J. An Evolutionary Approach to Multivariate Feature Selection for FMRI Pattern Analysis. BIOSIGNALS. 2008;(2):302–307. 2008.
-
- Chang C-C, Lin C-J. LIBSVM: a library for support vector machines, Software. 2001. available at http://www.csie.ntu.edu.tw~cjlin/libsvm.
-
- Cox DD, Savoy RL. Functional magnetic resonance imaging (fMRI) "brain reading": detecting and classifying distributed patterns of fMRI activity in human visual cortex. NeuroImage. 2003;19:261–270. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

