Analysis of complex single-molecule FRET time trajectories
- PMID: 20580964
- PMCID: PMC3012381
- DOI: 10.1016/S0076-6879(10)72011-5
Analysis of complex single-molecule FRET time trajectories
Abstract
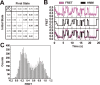
Single-molecule methods have given researchers the ability to investigate the structural dynamics of biomolecules at unprecedented resolution and sensitivity. One of the preferred methods of studying single biomolecules is single-molecule fluorescence resonance energy transfer (smFRET). The popularity of smFRET stems from its ability to report on dynamic, either intra- or intermolecular interactions in real-time. For example, smFRET has been successfully used to characterize the role of dynamics in functional RNAs and their protein complexes, including ribozymes, the ribosome, and more recently the spliceosome. Being able to reliably extract quantitative kinetic and conformational parameters from smFRET experiments is crucial for the interpretation of their results. The need for efficient, unbiased analysis routines becomes more evident as the systems studied become more complex. In this chapter, we focus on the practical utility of statistical algorithms, particularly hidden Markov models, to aid in the objective quantification of complex smFRET trajectories with three or more discrete states, and to extract kinetic information from the trajectories. Additionally, we present a method for systematically eliminating transitions associated with uncorrelated fluorophore behavior that may occur due to dye anisotropy and quenching effects. We also highlight the importance of data condensation through the use of various transition density plots to fully understand the underlying conformational dynamics and kinetic behavior of the biological macromolecule of interest under varying conditions. Finally, the application of these techniques to studies of pre-mRNA conformational changes during eukaryotic splicing is discussed.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures



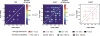
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

