TLR9 signaling is required for generation of the adaptive immune protection in Cryptococcus neoformans-infected lungs
- PMID: 20581055
- PMCID: PMC2913381
- DOI: 10.2353/ajpath.2010.091104
TLR9 signaling is required for generation of the adaptive immune protection in Cryptococcus neoformans-infected lungs
Abstract
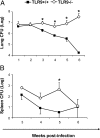
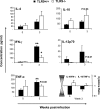
To determine whether TLR9 signaling contributes to the development of the adaptive immune response to cryptococcal infection, wild-type (TLR9+/+) and TLR9 knockout (TLR9-/-) BALB/c mice were infected intratracheally with 10(4) C. neoformans 52D. We evaluated 1) organ microbial burdens, 2) pulmonary leukocyte recruitment, 3) pulmonary and systemic cytokine induction, and 4) macrophage activation profiles. TLR9 deletion did not affect pulmonary growth during the innate phase, but profoundly impaired pulmonary clearance during the adaptive phase of the immune response (a 1000-fold difference at week 6). The impaired clearance in TLR9-/- mice was associated with: 1) significantly reduced CD4(+), CD8+ T cell, and CD19+ B cell recruitment into the lungs; 2) defects in Th polarization indicated by altered cytokine responses in the lungs, lymphonodes, and spleen; and 3) diminished macrophage accumulation and altered activation profile, including robust up-regulation of Arg1 and FIZZ1 (indicators of alternative activation) and diminished induction of inducible nitric oxide synthase (an indicator of classical activation). Histological analysis revealed defects in granuloma formation and increased numbers of intracellular yeast residing within macrophages in the lungs of TLR9-/- mice. We conclude that TLR9 signaling plays an important role in the development of robust protective immunity, proper recruitment and function of effector cells (lymphocytes and macrophages), and, ultimately, effective cryptococcal clearance from the infected lungs.
Figures






Similar articles
-
Early induction of CCL7 downstream of TLR9 signaling promotes the development of robust immunity to cryptococcal infection.J Immunol. 2012 Apr 15;188(8):3940-8. doi: 10.4049/jimmunol.1103053. Epub 2012 Mar 14. J Immunol. 2012. PMID: 22422883 Free PMC article.
-
Disruption of Early Tumor Necrosis Factor Alpha Signaling Prevents Classical Activation of Dendritic Cells in Lung-Associated Lymph Nodes and Development of Protective Immunity against Cryptococcal Infection.mBio. 2016 Jul 12;7(4):e00510-16. doi: 10.1128/mBio.00510-16. mBio. 2016. PMID: 27406560 Free PMC article.
-
STAT1 signaling is essential for protection against Cryptococcus neoformans infection in mice.J Immunol. 2014 Oct 15;193(8):4060-71. doi: 10.4049/jimmunol.1400318. Epub 2014 Sep 8. J Immunol. 2014. PMID: 25200956 Free PMC article.
-
Helper T-cell responses and pulmonary fungal infections.Immunology. 2018 Oct;155(2):155-163. doi: 10.1111/imm.12953. Epub 2018 Jun 14. Immunology. 2018. PMID: 29781185 Free PMC article. Review.
-
Androgen receptor influences on body defense system via modulation of innate and adaptive immune systems: lessons from conditional AR knockout mice.Am J Pathol. 2012 Nov;181(5):1504-12. doi: 10.1016/j.ajpath.2012.07.008. Epub 2012 Sep 4. Am J Pathol. 2012. PMID: 22959669 Free PMC article. Review.
Cited by
-
Role of dendritic cell-pathogen interactions in the immune response to pulmonary cryptococcal infection.Future Microbiol. 2015;10(11):1837-57. doi: 10.2217/fmb.15.92. Future Microbiol. 2015. PMID: 26597428 Free PMC article. Review.
-
Dendritic Cells: Multifunctional Roles in Host Defenses to Cryptococcus Infections.J Fungi (Basel). 2023 Oct 26;9(11):1050. doi: 10.3390/jof9111050. J Fungi (Basel). 2023. PMID: 37998856 Free PMC article. Review.
-
Intracellular Action of a Secreted Peptide Required for Fungal Virulence.Cell Host Microbe. 2016 Jun 8;19(6):849-64. doi: 10.1016/j.chom.2016.05.001. Epub 2016 May 19. Cell Host Microbe. 2016. PMID: 27212659 Free PMC article.
-
Pulmonary Macrophage and Dendritic Cell Responses to Cryptococcus neoformans.Front Cell Infect Microbiol. 2020 Feb 11;10:37. doi: 10.3389/fcimb.2020.00037. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32117810 Free PMC article. Review.
-
Toll like receptors in diseases of the lung.Int Immunopharmacol. 2011 Oct;11(10):1399-406. doi: 10.1016/j.intimp.2011.05.013. Epub 2011 May 30. Int Immunopharmacol. 2011. PMID: 21624505 Free PMC article. Review.
References
-
- Bottone EJ, Toma M, Johansson BE, Wormser GP. Capsule-deficient Cryptococcus neoformans in AIDS patients. Lancet. 1985;1:400. - PubMed
-
- Nishikawa MM, Lazera MS, Barbosa GG, Trilles L, Balassiano BR, Macedo RC, Bezerra CC, Perez MA, Cardarelli P, Wanke B. Serotyping of 467 Cryptococcus neoformans isolates from clinical and environmental sources in Brazil: analysis of host and regional patterns. J Clin Microbiol. 2003;41:73–77. - PMC - PubMed
-
- Perfect JR. Cryptococcus neoformans: a sugar-coated killer with designer genes. FEMS Immunol Med Microbiol. 2005;45:395–404. - PubMed
-
- Attal HC, Grover S, Bansal MP, Chaubey BS, Joglekar VK. Capsule deficient Cryptococcus neoformans an unusual clinical presentation. J Assoc Physicians India. 1983;31:49–51. - PubMed
-
- Bava J, Solari R, Isla G, Troncoso A. Atypical forms of Cryptococcus neoformans in CSF of an AIDS patient. J Infect Dev Ctries. 2008;2:403–405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

