All-codon scanning identifies p53 cancer rescue mutations
- PMID: 20581117
- PMCID: PMC2978351
- DOI: 10.1093/nar/gkq571
All-codon scanning identifies p53 cancer rescue mutations
Abstract
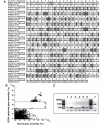
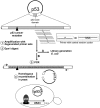
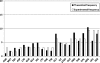
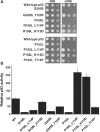
In vitro scanning mutagenesis strategies are valuable tools to identify critical residues in proteins and to generate proteins with modified properties. We describe the fast and simple All-Codon Scanning (ACS) strategy that creates a defined gene library wherein each individual codon within a specific target region is changed into all possible codons with only a single codon change per mutagenesis product. ACS is based on a multiplexed overlapping mutagenesis primer design that saturates only the targeted gene region with single codon changes. We have used ACS to produce single amino-acid changes in small and large regions of the human tumor suppressor protein p53 to identify single amino-acid substitutions that can restore activity to inactive p53 found in human cancers. Single-tube reactions were used to saturate defined 30-nt regions with all possible codon changes. The same technique was used in 20 parallel reactions to scan the 600-bp fragment encoding the entire p53 core domain. Identification of several novel p53 cancer rescue mutations demonstrated the utility of the ACS approach. ACS is a fast, simple and versatile method, which is useful for protein structure-function analyses and protein design or evolution problems.
Figures
References
-
- Jackel C, Kast P, Hilvert D. Protein design by directed evolution. Annu. Rev. Biophys. 2008;37:153–173. - PubMed
-
- Liu W, Sheppeck J.E., 2nd, Colby DA, Huang HB, Nairn AC, Chamberlin AR. The selective inhibition of phosphatases by natural toxins: the anhydride domain of tautomycin is not a primary factor in controlling PP1/PP2A selectivity. Bioorg. Med. Chem. Lett. 2003;13:1597–1600. - PubMed
-
- Hogrefe HH, Cline J, Youngblood GL, Allen RM. Creating randomized amino acid libraries with the quikchange multi site-directed mutagenesis kit. Biotechniques. 2002;33 1158–1160, 1162, 1164–1155. - PubMed
-
- Kegler-Ebo DM, Polack GW, DiMaio D. Use of codon cassette mutagenesis for saturation mutagenesis. Methods Mol. Biol. 1996;57:297–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

