Compartmentalized glucose metabolism in Pseudomonas putida is controlled by the PtxS repressor
- PMID: 20581202
- PMCID: PMC2937388
- DOI: 10.1128/JB.00520-10
Compartmentalized glucose metabolism in Pseudomonas putida is controlled by the PtxS repressor
Abstract
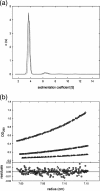
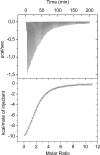
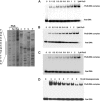
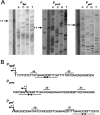
Metabolic flux analysis revealed that in Pseudomonas putida KT2440 about 50% of glucose taken up by the cells is channeled through the 2-ketogluconate peripheral pathway. This pathway is characterized by being compartmentalized in the cells. In fact, initial metabolism of glucose to 2-ketogluconate takes place in the periplasm through a set of reactions catalyzed by glucose dehydrogenase and gluconate dehydrogenase to yield 2-ketogluconate. This metabolite is subsequently transported to the cytoplasm, where two reactions are carried out, giving rise to 6-phosphogluconate, which enters the Entner-Doudoroff pathway. The genes for the periplasmic and cytoplasmic set of reactions are clustered in the host chromosome and grouped within two independent operons that are under the control of the PtxS regulator, which also modulates its own synthesis. Here, we show that although the two catabolic operons are induced in vivo by glucose, ketogluconate, and 2-ketogluconate, in vitro we found that only 2-ketogluconate binds to the regulator with an apparent K(D) (equilibrium dissociation constant) of 15 muM, as determined using isothermal titration calorimetry assays. PtxS is made of two domains, a helix-turn-helix DNA-binding domain located at the N terminus and a C-terminal domain that binds the effector. Differential scanning calorimetry assays revealed that PtxS unfolds via two events characterized by melting points of 48.1 degrees C and 57.6 degrees C and that, in the presence of 2-ketogluconate, the unfolding of the effector binding domain occurs at a higher temperature, providing further evidence for 2-ketogluconate-PtxS interactions. Purified PtxS is a dimer that binds to the target promoters with affinities in the range of 1 to 3 muM. Footprint analysis revealed that PtxS binds to an almost perfect palindrome that is present within the three promoters and whose consensus sequence is 5'-TGAAACCGGTTTCA-3'. This palindrome overlaps with the RNA polymerase binding site.
Figures




Similar articles
-
Genes for carbon metabolism and the ToxA virulence factor in Pseudomonas aeruginosa are regulated through molecular interactions of PtxR and PtxS.PLoS One. 2012;7(7):e39390. doi: 10.1371/journal.pone.0039390. Epub 2012 Jul 23. PLoS One. 2012. PMID: 22844393 Free PMC article.
-
Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis.J Bacteriol. 2007 Jul;189(14):5142-52. doi: 10.1128/JB.00203-07. Epub 2007 May 4. J Bacteriol. 2007. PMID: 17483213 Free PMC article.
-
A set of activators and repressors control peripheral glucose pathways in Pseudomonas putida to yield a common central intermediate.J Bacteriol. 2008 Apr;190(7):2331-9. doi: 10.1128/JB.01726-07. Epub 2008 Feb 1. J Bacteriol. 2008. PMID: 18245293 Free PMC article.
-
Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme.Gene. 1998 Nov 26;223(1-2):257-67. doi: 10.1016/s0378-1119(98)00366-7. Gene. 1998. PMID: 9858745 Review.
-
Transcriptional control of the Pseudomonas putida TOL plasmid catabolic pathways.Mol Microbiol. 1993 Sep;9(5):923-9. doi: 10.1111/j.1365-2958.1993.tb01222.x. Mol Microbiol. 1993. PMID: 7934920 Review.
Cited by
-
Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida.J Biol Chem. 2011 Mar 18;286(11):9351-9. doi: 10.1074/jbc.M110.187583. Epub 2011 Jan 14. J Biol Chem. 2011. PMID: 21239488 Free PMC article.
-
Molecular Characterization of a Transcriptional Regulator GntR for Gluconate Metabolism in Industrial 2-Ketogluconate Producer Pseudomonas plecoglossicida JUIM01.Microorganisms. 2025 Jun 15;13(6):1395. doi: 10.3390/microorganisms13061395. Microorganisms. 2025. PMID: 40572283 Free PMC article.
-
Gene expression reprogramming of Pseudomonas alloputida in response to arginine through the transcriptional regulator ArgR.Microbiology (Reading). 2024 Mar;170(3):001449. doi: 10.1099/mic.0.001449. Microbiology (Reading). 2024. PMID: 38511653 Free PMC article.
-
GtrS and GltR form a two-component system: the central role of 2-ketogluconate in the expression of exotoxin A and glucose catabolic enzymes in Pseudomonas aeruginosa.Nucleic Acids Res. 2014 Jul;42(12):7654-63. doi: 10.1093/nar/gku496. Epub 2014 Jun 11. Nucleic Acids Res. 2014. PMID: 24920832 Free PMC article.
-
A reduction in growth rate of Pseudomonas putida KT2442 counteracts productivity advances in medium-chain-length polyhydroxyalkanoate production from gluconate.Microb Cell Fact. 2011 Apr 22;10:25. doi: 10.1186/1475-2859-10-25. Microb Cell Fact. 2011. PMID: 21513516 Free PMC article.
References
-
- Colmer, J. A., and A. N. Hamood. 1998. Characterization of ptxS, a Pseudomonas aeruginosa gene which interferes with the effect of the exotoxin A positive regulatory gene, ptxR. Mol. Gen. Genet. 258:250-259. - PubMed
-
- Colmer-Hamood, J. A., H. Aramaki, J. M. Gaines, and A. N. Hamood. 2006. Transcriptional analysis of the Pseudomonas aeruginosa toxA regulatory gene ptxR. Can. J. Microbiol. 52:343-356. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

