Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces
- PMID: 20581307
- PMCID: PMC2910976
- DOI: 10.1105/tpc.110.074187
Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces
Abstract
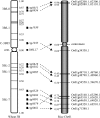
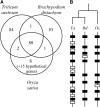
To improve our understanding of the organization and evolution of the wheat (Triticum aestivum) genome, we sequenced and annotated 13-Mb contigs (18.2 Mb) originating from different regions of its largest chromosome, 3B (1 Gb), and produced a 2x chromosome survey by shotgun Illumina/Solexa sequencing. All regions carried genes irrespective of their chromosomal location. However, gene distribution was not random, with 75% of them clustered into small islands containing three genes on average. A twofold increase of gene density was observed toward the telomeres likely due to high tandem and interchromosomal duplication events. A total of 3222 transposable elements were identified, including 800 new families. Most of them are complete but showed a highly nested structure spread over distances as large as 200 kb. A succession of amplification waves involving different transposable element families led to contrasted sequence compositions between the proximal and distal regions. Finally, with an estimate of 50,000 genes per diploid genome, our data suggest that wheat may have a higher gene number than other cereals. Indeed, comparisons with rice (Oryza sativa) and Brachypodium revealed that a high number of additional noncollinear genes are interspersed within a highly conserved ancestral grass gene backbone, supporting the idea of an accelerated evolution in the Triticeae lineages.
Figures



Comment in
-
On the habits of transposons: Dissociation mapping in maize and megabase sequencing in wheat reveal site preferences, distribution, and evolutionary history.Plant Cell. 2010 Jun;22(6):1650-2. doi: 10.1105/tpc.110.077396. Plant Cell. 2010. PMID: 20695122 Free PMC article. No abstract available.
References
-
- Akhunov E.D., Akhunova A.R., Dvorak J. (2007). Mechanisms and rates of birth and death of dispersed duplicated genes during the evolution of a multigene family in diploid and tetraploid wheats. Mol. Biol. Evol. 24: 539–550 - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

