Transcriptional regulation by small RNAs at sequences downstream from 3' gene termini
- PMID: 20581822
- PMCID: PMC3909968
- DOI: 10.1038/nchembio.400
Transcriptional regulation by small RNAs at sequences downstream from 3' gene termini
Abstract
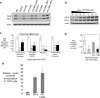
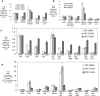
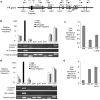
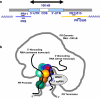
Transcriptome studies reveal many noncoding transcripts overlapping 3' gene termini. The function of these transcripts is unknown. Here we have characterized transcription at the progesterone receptor (PR) locus and identified noncoding transcripts that overlap the 3' end of the gene. Small RNAs complementary to sequences beyond the 3' terminus of PR mRNA modulated expression of PR, recruited argonaute 2 to a 3' noncoding transcript, altered occupancy of RNA polymerase II, induced chromatin changes at the PR promoter and affected responses to physiological stimuli. We found that the promoter and 3' terminal regions of the PR locus are in close proximity, providing a potential mechanism for RNA-mediated control of transcription over long genomic distances. These results extend the potential for small RNAs to regulate transcription to target sequences beyond the 3' termini of mRNA.
Figures





References
-
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. - PubMed
-
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92. - PubMed
-
- Janowski BA, et al. Inhibiting gene expression at transcription start sites in chromosomal DNA by antigene RNAs. Nat. Chem. Biol. 2005;1:216–222. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

