Structural and functional insights into 5'-ppp RNA pattern recognition by the innate immune receptor RIG-I
- PMID: 20581823
- PMCID: PMC3744876
- DOI: 10.1038/nsmb.1863
Structural and functional insights into 5'-ppp RNA pattern recognition by the innate immune receptor RIG-I
Abstract
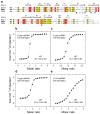
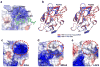
RIG-I is a cytosolic helicase that senses 5'-ppp RNA contained in negative-strand RNA viruses and triggers innate antiviral immune responses. Calorimetric binding studies established that the RIG-I C-terminal regulatory domain (CTD) binds to blunt-end double-stranded 5'-ppp RNA a factor of 17 more tightly than to its single-stranded counterpart. Here we report on the crystal structure of RIG-I CTD bound to both blunt ends of a self-complementary 5'-ppp dsRNA 12-mer, with interactions involving 5'-pp clearly visible in the complex. The structure, supported by mutation studies, defines how a lysine-rich basic cleft within the RIG-I CTD sequesters the observable 5'-pp of the bound RNA, with a stacked phenylalanine capping the terminal base pair. Key intermolecular interactions observed in the crystalline state are retained in the complex of 5'-ppp dsRNA 24-mer and full-length RIG-I under in vivo conditions, as evaluated from the impact of binding pocket RIG-I mutations and 2'-OCH(3) RNA modifications on the interferon response.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests. T.T. is a co-founder and scientific advisor of Alnylam Pharmaceuticals, and also an advisor of Regulus Therapeutics.
Figures




References
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. - PubMed
-
- Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev Med Virol. 2010;20:4–22. - PubMed
-
- Rehwinkel J, Reis e Sousa C. RIGorous detection: Exposing virus through RNA sensing. Science. 2010;327:284–286. - PubMed
-
- Coch C, et al. Higher activation of TLR9 in plasmacytoid dendritic cells by microbial DNA compared with self-DNA based on CpG-specific recognition of phosphodiester DNA. J Leukoc Biol. 2009;86:663–670. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

