Consistency of Random Survival Forests
- PMID: 20582150
- PMCID: PMC2889677
- DOI: 10.1016/j.spl.2010.02.020
Consistency of Random Survival Forests
Abstract
We prove uniform consistency of Random Survival Forests (RSF), a newly introduced forest ensemble learner for analysis of right-censored survival data. Consistency is proven under general splitting rules, bootstrapping, and random selection of variables-that is, under true implementation of the methodology. Under this setting we show that the forest ensemble survival function converges uniformly to the true population survival function. To prove this result we make one key assumption regarding the feature space: we assume that all variables are factors. Doing so ensures that the feature space has finite cardinality and enables us to exploit counting process theory and the uniform consistency of the Kaplan-Meier survival function.
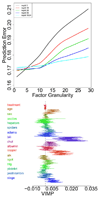
Figures

References
-
- Amit Y, Geman D. Shape quantization and recognition with randomized trees. Neural Computation. 1997;9:1545–1588.
-
- Andersen PK, Borgan O, Gill RD, Keiding N. Statistical Methods Based on Counting Processes. New York: Springer; 1993.
-
- Biau G, Devroye L, Lugosi G. Consistency of random forests and other classifiers. J. Machine Learning Research. 2008;9:2039–2057.
-
- Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression Trees. Belmont, California: 1984.
-
- Breiman L. Bagging predictors. Machine Learning. 1996;26:123–140.
Grants and funding
LinkOut - more resources
Full Text Sources
