Image-based models of cardiac structure in health and disease
- PMID: 20582162
- PMCID: PMC2889712
- DOI: 10.1002/wsbm.76
Image-based models of cardiac structure in health and disease
Abstract
Computational approaches to investigating the electromechanics of healthy and diseased hearts are becoming essential for the comprehensive understanding of cardiac function. In this article, we first present a brief review of existing image-based computational models of cardiac structure. We then provide a detailed explanation of a processing pipeline which we have recently developed for constructing realistic computational models of the heart from high resolution structural and diffusion tensor (DT) magnetic resonance (MR) images acquired ex vivo. The presentation of the pipeline incorporates a review of the methodologies that can be used to reconstruct models of cardiac structure. In this pipeline, the structural image is segmented to reconstruct the ventricles, normal myocardium, and infarct. A finite element mesh is generated from the segmented structural image, and fiber orientations are assigned to the elements based on DTMR data. The methods were applied to construct seven different models of healthy and diseased hearts. These models contain millions of elements, with spatial resolutions in the order of hundreds of microns, providing unprecedented detail in the representation of cardiac structure for simulation studies.
Figures
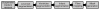




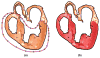
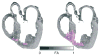


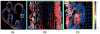
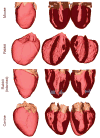
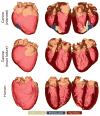
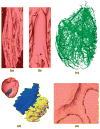

References
-
- Rodgers A, Vaughan P. The World Health Report 2002. Geneva, Switzerland: The World Health Organization; 2002.
-
- Noble D. Modeling the heart–from genes to cells to the whole organ. Science. 2002;295:1678–1682. - PubMed
-
- Hunter PJ, Pullan AJ, Smaill BH. Modeling total heart function. Ann Rev Biomed Eng. 2003;5:147–177. - PubMed
-
- Trayanova N, Eason J, Aguel F. Computer simulations of cardiac defibrillation: a look inside the heart. Comput Vis in Sci. 2002;4:259–270.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

