Data analysis in flow cytometry: the future just started
- PMID: 20583274
- PMCID: PMC2909632
- DOI: 10.1002/cyto.a.20901
Data analysis in flow cytometry: the future just started
Abstract
In the last 10 years, a tremendous progress characterized flow cytometry in its different aspects. In particular, major advances have been conducted regarding the hardware/instrumentation and reagent development, thus allowing fine cell analysis up to 20 parameters. As a result, this technology generates very complex datasets that demand for the development of optimal tools of analysis. Recently, many independent research groups approached the problem by using both supervised and unsupervised methods. In this article, we will review the new developments concerning the use of bioinformatics for polychromatic flow cytometry and propose what should be done to unravel the enormous heterogeneity of the cells we interrogate each day.
Figures
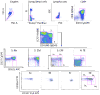
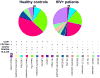
References
-
- De Rosa SC, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;7(2):245–8. - PubMed
-
- Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, Perfetto SP, Goepfert P, Koup RA, De Rosa SC, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12(8):972–7. - PubMed
-
- Suni MA, Picker LJ, Maino VC. Detection of antigen-specific T cell cytokine expression in whole blood by flow cytometry. J Immunol Methods. 1998;212(1):89–98. - PubMed
-
- Perez OD, Nolan GP. Simultaneous measurement of multiple active kinase states using polychromatic flow cytometry. Nat Biotechnol. 2002;20(2):155–62. - PubMed
-
- Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308(5721):523–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

