Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA
- PMID: 20584901
- PMCID: PMC2937867
- DOI: 10.1074/jbc.M110.106831
Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA
Abstract
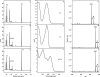
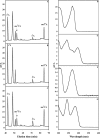
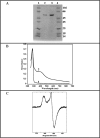
Bacterial and eukaryotic transfer RNAs have been shown to contain hypermodified adenosine, 2-methylthio-N(6)-threonylcarbamoyladenosine, at position 37 (A(37)) adjacent to the 3'-end of the anticodon, which is essential for efficient and highly accurate protein translation by the ribosome. Using a combination of bioinformatic sequence analysis and in vivo assay coupled to HPLC/MS technique, we have identified, from distinct sequence signatures, two methylthiotransferase (MTTase) subfamilies, designated as MtaB in bacterial cells and e-MtaB in eukaryotic and archaeal cells. Both subfamilies are responsible for the transformation of N(6)-threonylcarbamoyladenosine into 2-methylthio-N(6)-threonylcarbamoyladenosine. Recently, a variant within the human CDKAL1 gene belonging to the e-MtaB subfamily was shown to predispose for type 2 diabetes. CDKAL1 is thus the first eukaryotic MTTase identified so far. Using purified preparations of Bacillus subtilis MtaB (YqeV), a CDKAL1 bacterial homolog, we demonstrate that YqeV/CDKAL1 enzymes, as the previously studied MTTases MiaB and RimO, contain two [4Fe-4S] clusters. This work lays the foundation for elucidating the function of CDKAL1.
Figures

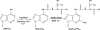

Similar articles
-
Functional characterization of the YmcB and YqeV tRNA methylthiotransferases of Bacillus subtilis.Nucleic Acids Res. 2010 Oct;38(18):6195-205. doi: 10.1093/nar/gkq364. Epub 2010 May 14. Nucleic Acids Res. 2010. PMID: 20472640 Free PMC article.
-
Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs.Nucleic Acids Res. 2017 Feb 28;45(4):2124-2136. doi: 10.1093/nar/gkw1120. Nucleic Acids Res. 2017. PMID: 27913733 Free PMC article.
-
MiaB protein from Thermotoga maritima. Characterization of an extremely thermophilic tRNA-methylthiotransferase.J Biol Chem. 2003 Aug 8;278(32):29515-24. doi: 10.1074/jbc.M301518200. Epub 2003 May 24. J Biol Chem. 2003. PMID: 12766153
-
Functional loss of Cdkal1, a novel tRNA modification enzyme, causes the development of type 2 diabetes.Endocr J. 2011;58(10):819-25. doi: 10.1507/endocrj.ej11-0099. Epub 2011 Sep 8. Endocr J. 2011. PMID: 21908934 Review.
-
The Biology of tRNA t6A Modification and Hypermodifications-Biogenesis and Disease Relevance.J Mol Biol. 2025 Aug 15;437(16):169091. doi: 10.1016/j.jmb.2025.169091. Epub 2025 Mar 27. J Mol Biol. 2025. PMID: 40155300 Review.
Cited by
-
Unanticipated coordination of tris buffer to the Radical SAM cluster of the RimO methylthiotransferase.J Biol Inorg Chem. 2016 Jul;21(4):549-57. doi: 10.1007/s00775-016-1365-8. Epub 2016 Jun 3. J Biol Inorg Chem. 2016. PMID: 27259294
-
The CDK5 repressor CDK5RAP1 is a methylthiotransferase acting on nuclear and mitochondrial RNA.Nucleic Acids Res. 2012 Jul;40(13):6235-40. doi: 10.1093/nar/gks240. Epub 2012 Mar 15. Nucleic Acids Res. 2012. PMID: 22422838 Free PMC article.
-
CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1) is a tail-anchored protein in the endoplasmic reticulum (ER) of insulinoma cells.J Biol Chem. 2012 Dec 7;287(50):41808-19. doi: 10.1074/jbc.M112.376558. Epub 2012 Oct 9. J Biol Chem. 2012. PMID: 23048041 Free PMC article.
-
Conservation and Diversification of tRNA t6A-Modifying Enzymes across the Three Domains of Life.Int J Mol Sci. 2022 Nov 6;23(21):13600. doi: 10.3390/ijms232113600. Int J Mol Sci. 2022. PMID: 36362385 Free PMC article. Review.
-
Impact of tRNA Modifications and tRNA-Modifying Enzymes on Proteostasis and Human Disease.Int J Mol Sci. 2018 Nov 24;19(12):3738. doi: 10.3390/ijms19123738. Int J Mol Sci. 2018. PMID: 30477220 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

