Genes of the ecdysone biosynthesis pathway are regulated by the dATAC histone acetyltransferase complex in Drosophila
- PMID: 20584983
- PMCID: PMC2937542
- DOI: 10.1128/MCB.00142-10
Genes of the ecdysone biosynthesis pathway are regulated by the dATAC histone acetyltransferase complex in Drosophila
Abstract
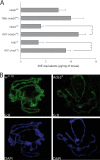
Uncovering mechanisms that regulate ecdysone production is an important step toward understanding the regulation of insect metamorphosis and processes in steroid-related pathologies. We report here the transcriptome analysis of Drosophila melanogaster dAda2a and dAda3 mutants, in which subunits of the ATAC acetyltransferase complex are affected. In agreement with the fact that these mutations lead to lethality at the start of metamorphosis, both the ecdysone levels and the ecdysone receptor binding to polytene chromosomes are reduced in these flies. The cytochrome genes (spookier, phantom, disembodied, and shadow) involved in steroid conversion in the ring gland are downregulated, while the gene shade, which is involved in converting ecdysone into its active form in the periphery, is upregulated in these dATAC subunit mutants. Moreover, driven expression of dAda3 at the site of ecdysone synthesis partially rescues dAda3 mutants. Mutants of dAda2b, a subunit of the dSAGA histone acetyltransferase complex, do not share phenotype characteristics and RNA profile alterations with dAda2a mutants, indicating that the ecdysone biosynthesis genes are regulated by dATAC, but not by dSAGA. Thus, we provide one of the first examples of the coordinated regulation of a functionally linked set of genes by the metazoan-specific ATAC complex.
Figures



Similar articles
-
dTAF10- and dTAF10b-Containing Complexes Are Required for Ecdysone-Driven Larval-Pupal Morphogenesis in Drosophila melanogaster.PLoS One. 2015 Nov 10;10(11):e0142226. doi: 10.1371/journal.pone.0142226. eCollection 2015. PLoS One. 2015. PMID: 26556600 Free PMC article.
-
Acetylations of Ftz-F1 and histone H4K5 are required for the fine-tuning of ecdysone biosynthesis during Drosophila metamorphosis.Dev Biol. 2015 Aug 1;404(1):80-7. doi: 10.1016/j.ydbio.2015.04.020. Epub 2015 May 8. Dev Biol. 2015. PMID: 25959239
-
GAL4 induces transcriptionally active puff in the absence of dSAGA- and ATAC-specific chromatin acetylation in the Drosophila melanogaster polytene chromosome.Chromosoma. 2009 Aug;118(4):513-26. doi: 10.1007/s00412-009-0215-7. Epub 2009 May 2. Chromosoma. 2009. PMID: 19412618
-
What goes up must come down: transcription factors have their say in making ecdysone pulses.Curr Top Dev Biol. 2013;103:35-71. doi: 10.1016/B978-0-12-385979-2.00002-2. Curr Top Dev Biol. 2013. PMID: 23347515 Review.
-
Ecdysone signaling cascade and regulation of Drosophila metamorphosis.Arch Insect Biochem Physiol. 1996;33(3-4):231-44. doi: 10.1002/(SICI)1520-6327(1996)33:3/4<231::AID-ARCH5>3.0.CO;2-V. Arch Insect Biochem Physiol. 1996. PMID: 8913033 Review.
Cited by
-
De novo characterization of Phenacoccus solenopsis transcriptome and analysis of gene expression profiling during development and hormone biosynthesis.Sci Rep. 2018 May 15;8(1):7573. doi: 10.1038/s41598-018-25845-3. Sci Rep. 2018. PMID: 29765069 Free PMC article.
-
Transcriptional regulation of insect steroid hormone biosynthesis and its role in controlling timing of molting and metamorphosis.Dev Growth Differ. 2016 Jan;58(1):94-105. doi: 10.1111/dgd.12248. Epub 2015 Dec 15. Dev Growth Differ. 2016. PMID: 26667894 Free PMC article. Review.
-
The Putzig-NURF nucleosome remodeling complex is required for ecdysone receptor signaling and innate immunity in Drosophila melanogaster.Genetics. 2011 May;188(1):127-39. doi: 10.1534/genetics.111.127795. Epub 2011 Mar 8. Genetics. 2011. PMID: 21385730 Free PMC article.
-
ATAC-king the complexity of SAGA during evolution.Genes Dev. 2012 Mar 15;26(6):527-41. doi: 10.1101/gad.184705.111. Genes Dev. 2012. PMID: 22426530 Free PMC article. Review.
-
An Overview of Embryogenesis: External Morphology and Transcriptome Profiling in the Hemipteran Insect Nilaparvata lugens.Front Physiol. 2020 Feb 18;11:106. doi: 10.3389/fphys.2020.00106. eCollection 2020. Front Physiol. 2020. PMID: 32132932 Free PMC article.
References
-
- Baehrecke, E. H. 1996. Ecdysone signaling cascade and regulation of Drosophila metamorphosis. Arch. Insect Biochem. Physiol. 33:231-244. - PubMed
-
- Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. - PubMed
-
- Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447:407-412. - PubMed
-
- Carré, C., A. Ciurciu, O. Komonyi, C. Jacquier, D. Fagegaltier, J. Pidoux, H. Tricoire, L. Tora, I. M. Boros, and C. Antoniewski. 2008. The Drosophila NURF remodelling and the ATAC histone acetylase complexes functionally interact and are required for global chromosome organization. EMBO Rep. 9:187-192. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
