NeuroML: a language for describing data driven models of neurons and networks with a high degree of biological detail
- PMID: 20585541
- PMCID: PMC2887454
- DOI: 10.1371/journal.pcbi.1000815
NeuroML: a language for describing data driven models of neurons and networks with a high degree of biological detail
Abstract
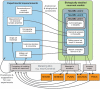
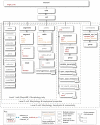
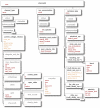
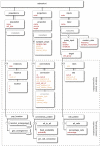
Biologically detailed single neuron and network models are important for understanding how ion channels, synapses and anatomical connectivity underlie the complex electrical behavior of the brain. While neuronal simulators such as NEURON, GENESIS, MOOSE, NEST, and PSICS facilitate the development of these data-driven neuronal models, the specialized languages they employ are generally not interoperable, limiting model accessibility and preventing reuse of model components and cross-simulator validation. To overcome these problems we have used an Open Source software approach to develop NeuroML, a neuronal model description language based on XML (Extensible Markup Language). This enables these detailed models and their components to be defined in a standalone form, allowing them to be used across multiple simulators and archived in a standardized format. Here we describe the structure of NeuroML and demonstrate its scope by converting into NeuroML models of a number of different voltage- and ligand-gated conductances, models of electrical coupling, synaptic transmission and short-term plasticity, together with morphologically detailed models of individual neurons. We have also used these NeuroML-based components to develop an highly detailed cortical network model. NeuroML-based model descriptions were validated by demonstrating similar model behavior across five independently developed simulators. Although our results confirm that simulations run on different simulators converge, they reveal limits to model interoperability, by showing that for some models convergence only occurs at high levels of spatial and temporal discretisation, when the computational overhead is high. Our development of NeuroML as a common description language for biophysically detailed neuronal and network models enables interoperability across multiple simulation environments, thereby improving model transparency, accessibility and reuse in computational neuroscience.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
The NeuroML ecosystem for standardized multi-scale modeling in neuroscience.Elife. 2025 Jan 10;13:RP95135. doi: 10.7554/eLife.95135. Elife. 2025. PMID: 39792574 Free PMC article.
-
NeuroML-DB: Sharing and characterizing data-driven neuroscience models described in NeuroML.PLoS Comput Biol. 2023 Mar 3;19(3):e1010941. doi: 10.1371/journal.pcbi.1010941. eCollection 2023 Mar. PLoS Comput Biol. 2023. PMID: 36867658 Free PMC article.
-
EDEN: A High-Performance, General-Purpose, NeuroML-Based Neural Simulator.Front Neuroinform. 2022 May 20;16:724336. doi: 10.3389/fninf.2022.724336. eCollection 2022. Front Neuroinform. 2022. PMID: 35669596 Free PMC article.
-
MorphML: level 1 of the NeuroML standards for neuronal morphology data and model specification.Neuroinformatics. 2007 Summer;5(2):96-104. doi: 10.1007/s12021-007-0003-6. Neuroinformatics. 2007. PMID: 17873371 Free PMC article. Review.
-
Towards NeuroML: model description methods for collaborative modelling in neuroscience.Philos Trans R Soc Lond B Biol Sci. 2001 Aug 29;356(1412):1209-28. doi: 10.1098/rstb.2001.0910. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11545699 Free PMC article. Review.
Cited by
-
SpineCreator: a Graphical User Interface for the Creation of Layered Neural Models.Neuroinformatics. 2017 Jan;15(1):25-40. doi: 10.1007/s12021-016-9311-z. Neuroinformatics. 2017. PMID: 27628934 Free PMC article.
-
Reproducibility in Computational Neuroscience Models and Simulations.IEEE Trans Biomed Eng. 2016 Oct;63(10):2021-35. doi: 10.1109/TBME.2016.2539602. Epub 2016 Mar 8. IEEE Trans Biomed Eng. 2016. PMID: 27046845 Free PMC article.
-
Modeling the Cerebellar Microcircuit: New Strategies for a Long-Standing Issue.Front Cell Neurosci. 2016 Jul 8;10:176. doi: 10.3389/fncel.2016.00176. eCollection 2016. Front Cell Neurosci. 2016. PMID: 27458345 Free PMC article. Review.
-
The first 10 years of the international coordination network for standards in systems and synthetic biology (COMBINE).J Integr Bioinform. 2020 Jun 29;17(2-3):20200005. doi: 10.1515/jib-2020-0005. J Integr Bioinform. 2020. PMID: 32598315 Free PMC article.
-
c302: a multiscale framework for modelling the nervous system of Caenorhabditis elegans.Philos Trans R Soc Lond B Biol Sci. 2018 Sep 10;373(1758):20170379. doi: 10.1098/rstb.2017.0379. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 30201842 Free PMC article.
References
-
- Mainen ZF, Sejnowski TJ. Influence of dendritic structure on firing pattern in model neocortical neurons. Nature. 1996;382:363–366. - PubMed
-
- Kole MH, Ilschner SU, Kampa BM, Williams SR, Ruben PC, et al. Action potential generation requires a high sodium channel density in the axon initial segment. Nat Neurosci. 2008;11:178–186. - PubMed
-
- Jarsky T, Roxin A, Kath WL, Spruston N. Conditional dendritic spike propagation following distal synaptic activation of hippocampal CA1 pyramidal neurons. Nat Neurosci. 2005;8:1667–1676. - PubMed
Publication types
MeSH terms
Grants and funding
- 086699/WT_/Wellcome Trust/United Kingdom
- R01 NS011613/NS/NINDS NIH HHS/United States
- R01 MH081905/MH/NIMH NIH HHS/United States
- P01 DC04732/DC/NIDCD NIH HHS/United States
- R01 DC009977/DC/NIDCD NIH HHS/United States
- R01 MH086638/MH/NIMH NIH HHS/United States
- R01 NS11613/NS/NINDS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- BB/F005490/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0400598/MRC_/Medical Research Council/United Kingdom
- P01 DC004732/DC/NIDCD NIH HHS/United States
- 0064413/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

