Revisiting date and party hubs: novel approaches to role assignment in protein interaction networks
- PMID: 20585543
- PMCID: PMC2887459
- DOI: 10.1371/journal.pcbi.1000817
Revisiting date and party hubs: novel approaches to role assignment in protein interaction networks
Abstract
The idea of "date" and "party" hubs has been influential in the study of protein-protein interaction networks. Date hubs display low co-expression with their partners, whilst party hubs have high co-expression. It was proposed that party hubs are local coordinators whereas date hubs are global connectors. Here, we show that the reported importance of date hubs to network connectivity can in fact be attributed to a tiny subset of them. Crucially, these few, extremely central, hubs do not display particularly low expression correlation, undermining the idea of a link between this quantity and hub function. The date/party distinction was originally motivated by an approximately bimodal distribution of hub co-expression; we show that this feature is not always robust to methodological changes. Additionally, topological properties of hubs do not in general correlate with co-expression. However, we find significant correlations between interaction centrality and the functional similarity of the interacting proteins. We suggest that thinking in terms of a date/party dichotomy for hubs in protein interaction networks is not meaningful, and it might be more useful to conceive of roles for protein-protein interactions rather than for individual proteins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
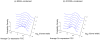
 -value
-value  ) for (a) and 0.0062 (
) for (a) and 0.0062 ( -value
-value  ) for (b), indicating no significant deviation from unimodality in either case.
) for (b), indicating no significant deviation from unimodality in either case.
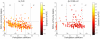
 (
( -score
-score  ,
,  -value
-value  ) for FYI,
) for FYI,  (
( -score
-score  ,
,  -value
-value  ) for FHC.
) for FHC.References
-
- Han JDJ, Bertin N, Hao T, Goldberg DS, Berriz GF, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. - PubMed
-
- Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nature Biotechnology. 2009;27:199–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

