Beauty is in the eye of the beholder: proteins can recognize binding sites of homologous proteins in more than one way
- PMID: 20585553
- PMCID: PMC2887470
- DOI: 10.1371/journal.pcbi.1000821
Beauty is in the eye of the beholder: proteins can recognize binding sites of homologous proteins in more than one way
Abstract
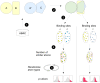
Understanding the mechanisms of protein-protein interaction is a fundamental problem with many practical applications. The fact that different proteins can bind similar partners suggests that convergently evolved binding interfaces are reused in different complexes. A set of protein complexes composed of non-homologous domains interacting with homologous partners at equivalent binding sites was collected in 2006, offering an opportunity to investigate this point. We considered 433 pairs of protein-protein complexes from the ABAC database (AB and AC binary protein complexes sharing a homologous partner A) and analyzed the extent of physico-chemical similarity at the atomic and residue level at the protein-protein interface. Homologous partners of the complexes were superimposed using Multiprot, and similar atoms at the interface were quantified using a five class grouping scheme and a distance cut-off. We found that the number of interfacial atoms with similar properties is systematically lower in the non-homologous proteins than in the homologous ones. We assessed the significance of the similarity by bootstrapping the atomic properties at the interfaces. We found that the similarity of binding sites is very significant between homologous proteins, as expected, but generally insignificant between the non-homologous proteins that bind to homologous partners. Furthermore, evolutionarily conserved residues are not colocalized within the binding sites of non-homologous proteins. We could only identify a limited number of cases of structural mimicry at the interface, suggesting that this property is less generic than previously thought. Our results support the hypothesis that different proteins can interact with similar partners using alternate strategies, but do not support convergent evolution.
Conflict of interest statement
The author has declared that no competing interests exist.
Figures
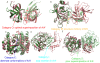
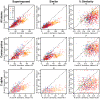
 representations. First column: number of superimposed elements on A/A′ versus B/C side, second column: number of similar elements on A/A′ versus B/C side, third column: fraction of similar elements on A/A′ versus B/C side.
representations. First column: number of superimposed elements on A/A′ versus B/C side, second column: number of similar elements on A/A′ versus B/C side, third column: fraction of similar elements on A/A′ versus B/C side.
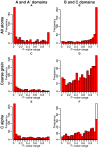
 representations, first column: P-values of the A/A′ domains, second column: P-values of the B/C domains. White bars correspond to a number of similar elements equal to zero, which, by definition, yields a P-value equal to 1, since the random model cannot give a number of similar residues lower than zero.
representations, first column: P-values of the A/A′ domains, second column: P-values of the B/C domains. White bars correspond to a number of similar elements equal to zero, which, by definition, yields a P-value equal to 1, since the random model cannot give a number of similar residues lower than zero.
Similar articles
-
Equivalent binding sites reveal convergently evolved interaction motifs.Bioinformatics. 2006 Mar 1;22(5):550-5. doi: 10.1093/bioinformatics/bti782. Epub 2005 Nov 15. Bioinformatics. 2006. PMID: 16287935
-
3D-partner: a web server to infer interacting partners and binding models.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W561-7. doi: 10.1093/nar/gkm346. Epub 2007 May 21. Nucleic Acids Res. 2007. PMID: 17517763 Free PMC article.
-
Oligomerisation status and evolutionary conservation of interfaces of protein structural domain superfamilies.Mol Biosyst. 2013 Jul;9(7):1652-61. doi: 10.1039/c3mb25484d. Epub 2013 Mar 27. Mol Biosyst. 2013. PMID: 23532342
-
Protein interactions in 3D: from interface evolution to drug discovery.J Struct Biol. 2012 Sep;179(3):347-58. doi: 10.1016/j.jsb.2012.04.009. Epub 2012 May 1. J Struct Biol. 2012. PMID: 22595401 Review.
-
Review and comparative assessment of sequence-based predictors of protein-binding residues.Brief Bioinform. 2018 Sep 28;19(5):821-837. doi: 10.1093/bib/bbx022. Brief Bioinform. 2018. PMID: 28334258 Review.
Cited by
-
Role of β-lactamase residues in a common interface for binding the structurally unrelated inhibitory proteins BLIP and BLIP-II.Protein Sci. 2014 Sep;23(9):1235-46. doi: 10.1002/pro.2505. Epub 2014 Jul 1. Protein Sci. 2014. PMID: 24947275 Free PMC article.
-
Pocket-based drug design: exploring pocket space.AAPS J. 2013 Jan;15(1):228-41. doi: 10.1208/s12248-012-9426-6. Epub 2012 Nov 22. AAPS J. 2013. PMID: 23180158 Free PMC article. Review.
-
Progress and challenges in predicting protein interfaces.Brief Bioinform. 2016 Jan;17(1):117-31. doi: 10.1093/bib/bbv027. Epub 2015 May 13. Brief Bioinform. 2016. PMID: 25971595 Free PMC article. Review.
-
Protein binding specificity versus promiscuity.Curr Opin Struct Biol. 2011 Feb;21(1):50-61. doi: 10.1016/j.sbi.2010.10.002. Epub 2010 Nov 9. Curr Opin Struct Biol. 2011. PMID: 21071205 Free PMC article. Review.
-
HotRegion: a database of predicted hot spot clusters.Nucleic Acids Res. 2012 Jan;40(Database issue):D829-33. doi: 10.1093/nar/gkr929. Epub 2011 Nov 12. Nucleic Acids Res. 2012. PMID: 22080558 Free PMC article.
References
-
- Ruffner H, Bauer A, Bouwmeester T. Human protein-protein interaction networks and the value for drug discovery. Drug Discov Today. 2007;12:709–716. - PubMed
-
- Chothia C, Janin J. Principles of protein-protein recognition. Nature. 1975;256:705–708. - PubMed
-
- Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein-protein interfaces. J Mol Biol. 2004;336:943–955. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

