NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases
- PMID: 20585566
- PMCID: PMC2891834
- DOI: 10.1371/journal.ppat.1000960
NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases
Abstract
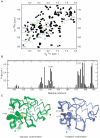
NleG homologues constitute the largest family of Type 3 effectors delivered by pathogenic E. coli, with fourteen members in the enterohaemorrhagic (EHEC) O157:H7 strain alone. Identified recently as part of the non-LEE-encoded (Nle) effector set, this family remained uncharacterised and shared no sequence homology to other proteins including those of known function. The C-terminal domain of NleG2-3 (residues 90 to 191) is the most conserved region in NleG proteins and was solved by NMR. Structural analysis of this structure revealed the presence of a RING finger/U-box motif. Functional assays demonstrated that NleG2-3 as well as NleG5-1, NleG6-2 and NleG9' family members exhibited a strong autoubiquitination activity in vitro; a characteristic usually expressed by eukaryotic ubiquitin E3 ligases. When screened for activity against a panel of 30 human E2 enzymes, the NleG2-3 and NleG5-1 homologues showed an identical profile with only UBE2E2, UBE2E3 and UBE2D2 enzymes supporting NleG activity. Fluorescence polarization analysis yielded a binding affinity constant of 56+/-2 microM for the UBE2D2/NleG5-1 interaction, a value comparable with previous studies on E2/E3 affinities. The UBE2D2 interaction interface on NleG2-3 defined by NMR chemical shift perturbation and mutagenesis was shown to be generally similar to that characterised for human RING finger ubiquitin ligases. The alanine substitutions of UBE2D2 residues Arg5 and Lys63, critical for activation of eukaryotic E3 ligases, also significantly decreased both NleG binding and autoubiquitination activity. These results demonstrate that bacteria-encoded NleG effectors are E3 ubiquitin ligases analogous to RING finger and U-box enzymes in eukaryotes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

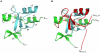




Similar articles
-
Functional diversification of the NleG effector family in enterohemorrhagic Escherichia coli.Proc Natl Acad Sci U S A. 2018 Oct 2;115(40):10004-10009. doi: 10.1073/pnas.1718350115. Epub 2018 Sep 14. Proc Natl Acad Sci U S A. 2018. PMID: 30217892 Free PMC article.
-
Mulan E3 ubiquitin ligase interacts with multiple E2 conjugating enzymes and participates in mitophagy by recruiting GABARAP.Cell Signal. 2014 Dec;26(12):2921-9. doi: 10.1016/j.cellsig.2014.09.004. Epub 2014 Sep 16. Cell Signal. 2014. PMID: 25224329
-
Distinct Molecular Features of NleG Type 3 Secreted Effectors Allow for Different Roles during Citrobacter rodentium Infection in Mice.Infect Immun. 2023 Jan 24;91(1):e0050522. doi: 10.1128/iai.00505-22. Epub 2022 Dec 13. Infect Immun. 2023. PMID: 36511702 Free PMC article.
-
The prolific ATL family of RING-H2 ubiquitin ligases.Plant Signal Behav. 2012 Aug;7(8):1014-21. doi: 10.4161/psb.20851. Epub 2012 Jul 25. Plant Signal Behav. 2012. PMID: 22827943 Free PMC article. Review.
-
TRIM proteins as RING finger E3 ubiquitin ligases.Adv Exp Med Biol. 2012;770:27-37. doi: 10.1007/978-1-4614-5398-7_3. Adv Exp Med Biol. 2012. PMID: 23630998 Review.
Cited by
-
TcpC inhibits toll-like receptor signaling pathway by serving as an E3 ubiquitin ligase that promotes degradation of myeloid differentiation factor 88.PLoS Pathog. 2021 Mar 31;17(3):e1009481. doi: 10.1371/journal.ppat.1009481. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33788895 Free PMC article.
-
The S. Typhi effector StoD is an E3/E4 ubiquitin ligase which binds K48- and K63-linked diubiquitin.Life Sci Alliance. 2019 May 29;2(3):e201800272. doi: 10.26508/lsa.201800272. Print 2019 Jun. Life Sci Alliance. 2019. PMID: 31142637 Free PMC article.
-
Effector triggered manipulation of host immune response elicited by different pathotypes of Escherichia coli.Virulence. 2014;5(7):733-9. doi: 10.4161/viru.29948. Virulence. 2014. PMID: 25513774 Free PMC article. Review.
-
Prevalence and Genomic Characterization of Escherichia coli O157:H7 in Cow-Calf Herds throughout California.Appl Environ Microbiol. 2017 Aug 1;83(16):e00734-17. doi: 10.1128/AEM.00734-17. Print 2017 Aug 15. Appl Environ Microbiol. 2017. PMID: 28550057 Free PMC article.
-
Exploitation of the host cell ubiquitin machinery by microbial effector proteins.J Cell Sci. 2017 Jun 15;130(12):1985-1996. doi: 10.1242/jcs.188482. Epub 2017 May 5. J Cell Sci. 2017. PMID: 28476939 Free PMC article. Review.
References
-
- Parsot C. Shigella type III secretion effectors: how, where, when, for what purposes? Curr Opin Microbiol. 2009;12:110–116. - PubMed
-
- Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–834. - PubMed
-
- Sal-Man N, Biemans-Oldehinkel E, Finlay BB. Structural microengineers: pathogenic Escherichia coli redesigns the actin cytoskeleton in host cells. Structure. 2009;17:15–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

