Human cytomegalovirus UL29/28 protein interacts with components of the NuRD complex which promote accumulation of immediate-early RNA
- PMID: 20585571
- PMCID: PMC2891856
- DOI: 10.1371/journal.ppat.1000965
Human cytomegalovirus UL29/28 protein interacts with components of the NuRD complex which promote accumulation of immediate-early RNA
Abstract
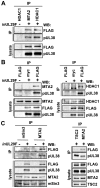
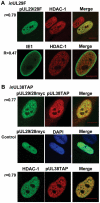
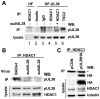
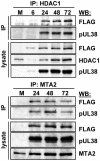
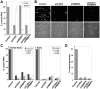
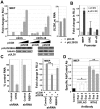
Histone deacetylation plays a pivotal role in regulating human cytomegalovirus gene expression. In this report, we have identified candidate HDAC1-interacting proteins in the context of infection by using a method for rapid immunoisolation of an epitope-tagged protein coupled with mass spectrometry. Putative interactors included multiple human cytomegalovirus-coded proteins. In particular, the interaction of pUL38 and pUL29/28 with HDAC1 was confirmed by reciprocal immunoprecipitations. HDAC1 is present in numerous protein complexes, including the HDAC1-containing nucleosome remodeling and deacetylase protein complex, NuRD. pUL38 and pUL29/28 associated with the MTA2 component of NuRD, and shRNA-mediated knockdown of the RBBP4 and CHD4 constituents of NuRD inhibited HCMV immediate-early RNA and viral DNA accumulation; together this argues that multiple components of the NuRD complex are needed for efficient HCMV replication. Consistent with a positive acting role for the NuRD elements during viral replication, the growth of pUL29/28- or pUL38-deficient viruses could not be rescued by treating infected cells with the deacetylase inhibitor, trichostatin A. Transient expression of pUL29/28 enhanced activity of the HCMV major immediate-early promoter in a reporter assay, regardless of pUL38 expression. Importantly, induction of the major immediate-early reporter activity by pUL29/28 required functional NuRD components, consistent with the inhibition of immediate-early RNA accumulation within infected cells after knockdown of RBBP4 and CHD4. We propose that pUL29/28 modifies the NuRD complex to stimulate the accumulation of immediate-early RNAs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Human cytomegalovirus pUL29/28 and pUL38 repression of p53-regulated p21CIP1 and caspase 1 promoters during infection.J Virol. 2013 Mar;87(5):2463-74. doi: 10.1128/JVI.01926-12. Epub 2012 Dec 12. J Virol. 2013. PMID: 23236067 Free PMC article.
-
Functional interaction of the human cytomegalovirus IE2 protein with histone deacetylase 2 in infected human fibroblasts.J Gen Virol. 2007 Dec;88(Pt 12):3214-3223. doi: 10.1099/vir.0.83171-0. J Gen Virol. 2007. PMID: 18024889
-
CHD4 Is a Peripheral Component of the Nucleosome Remodeling and Deacetylase Complex.J Biol Chem. 2016 Jul 22;291(30):15853-66. doi: 10.1074/jbc.M115.707018. Epub 2016 May 27. J Biol Chem. 2016. PMID: 27235397 Free PMC article.
-
Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics.FEBS J. 2015 May;282(9):1774-85. doi: 10.1111/febs.12972. Epub 2014 Sep 11. FEBS J. 2015. PMID: 25123934 Review.
-
The human Mi-2/NuRD complex and gene regulation.Oncogene. 2007 Aug 13;26(37):5433-8. doi: 10.1038/sj.onc.1210611. Oncogene. 2007. PMID: 17694084 Review.
Cited by
-
Human cytomegalovirus pUL97 regulates the viral major immediate early promoter by phosphorylation-mediated disruption of histone deacetylase 1 binding.J Virol. 2013 Jul;87(13):7393-408. doi: 10.1128/JVI.02825-12. Epub 2013 Apr 24. J Virol. 2013. PMID: 23616659 Free PMC article.
-
Mass spectrometry-based proteomic approaches for discovery of HIV-host interactions.Future Virol. 2014;9(11):979-992. doi: 10.2217/fvl.14.86. Future Virol. 2014. PMID: 25544858 Free PMC article.
-
Human cytomegalovirus IE1 protein disrupts interleukin-6 signaling by sequestering STAT3 in the nucleus.J Virol. 2013 Oct;87(19):10763-76. doi: 10.1128/JVI.01197-13. Epub 2013 Jul 31. J Virol. 2013. PMID: 23903834 Free PMC article.
-
The Impact of Mass Spectrometry-Based Proteomics on Fundamental Discoveries in Virology.Annu Rev Virol. 2014 Nov;1(1):581-604. doi: 10.1146/annurev-virology-031413-085527. Epub 2014 Jul 14. Annu Rev Virol. 2014. PMID: 26958735 Free PMC article.
-
Proteomic approaches to uncovering virus-host protein interactions during the progression of viral infection.Expert Rev Proteomics. 2016;13(3):325-40. doi: 10.1586/14789450.2016.1147353. Expert Rev Proteomics. 2016. PMID: 26817613 Free PMC article. Review.
References
-
- Mocarski E, Shenk T, Pass RF. Fields Virology: Lippincott Williams & Wilkins; 2007. Cytomegaloviruses. pp. 2702–2772.
-
- Waller EC, Day E, Sissons JG, Wills MR. Dynamics of T cell memory in human cytomegalovirus infection. Med Microbiol Immunol. 2008;197:83–96. - PubMed
-
- Cinatl J, Jr, Nevels M, Paulus C, Michaelis M. Activation of telomerase in glioma cells by human cytomegalovirus: another piece of the puzzle. J Natl Cancer Inst. 2009;101:441–443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DP1DA026192/DA/NIDA NIH HHS/United States
- CA85786/CA/NCI NIH HHS/United States
- R33 CA089810/CA/NCI NIH HHS/United States
- P41 RR000862/RR/NCRR NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- R01 CA085786/CA/NCI NIH HHS/United States
- CA89810/CA/NCI NIH HHS/United States
- RR00862/RR/NCRR NIH HHS/United States
- R01 GM062427/GM/NIGMS NIH HHS/United States
- DP1 DA026192/DA/NIDA NIH HHS/United States
- RR22220/RR/NCRR NIH HHS/United States
- R01 CA082396/CA/NCI NIH HHS/United States
- GM62427/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

