Drosophila genome-wide RNAi screen identifies multiple regulators of HIF-dependent transcription in hypoxia
- PMID: 20585616
- PMCID: PMC2891703
- DOI: 10.1371/journal.pgen.1000994
Drosophila genome-wide RNAi screen identifies multiple regulators of HIF-dependent transcription in hypoxia
Abstract
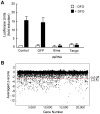
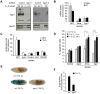
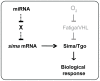
Hypoxia-inducible factors (HIFs) are a family of evolutionary conserved alpha-beta heterodimeric transcription factors that induce a wide range of genes in response to low oxygen tension. Molecular mechanisms that mediate oxygen-dependent HIF regulation operate at the level of the alpha subunit, controlling protein stability, subcellular localization, and transcriptional coactivator recruitment. We have conducted an unbiased genome-wide RNA interference (RNAi) screen in Drosophila cells aimed to the identification of genes required for HIF activity. After 3 rounds of selection, 30 genes emerged as critical HIF regulators in hypoxia, most of which had not been previously associated with HIF biology. The list of genes includes components of chromatin remodeling complexes, transcription elongation factors, and translational regulators. One remarkable hit was the argonaute 1 (ago1) gene, a central element of the microRNA (miRNA) translational silencing machinery. Further studies confirmed the physiological role of the miRNA machinery in HIF-dependent transcription. This study reveals the occurrence of novel mechanisms of HIF regulation, which might contribute to developing novel strategies for therapeutic intervention of HIF-related pathologies, including heart attack, cancer, and stroke.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
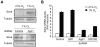

Similar articles
-
miRNAs regulate the HIF switch during hypoxia: a novel therapeutic target.Angiogenesis. 2018 May;21(2):183-202. doi: 10.1007/s10456-018-9600-2. Epub 2018 Jan 27. Angiogenesis. 2018. PMID: 29383635 Free PMC article. Review.
-
Hypoxia-inducible factor 3 biology: complexities and emerging themes.Am J Physiol Cell Physiol. 2016 Feb 15;310(4):C260-9. doi: 10.1152/ajpcell.00315.2015. Epub 2015 Nov 11. Am J Physiol Cell Physiol. 2016. PMID: 26561641 Review.
-
The insulin-PI3K/TOR pathway induces a HIF-dependent transcriptional response in Drosophila by promoting nuclear localization of HIF-alpha/Sima.J Cell Sci. 2005 Dec 1;118(Pt 23):5431-41. doi: 10.1242/jcs.02648. Epub 2005 Nov 8. J Cell Sci. 2005. PMID: 16278294
-
Progress on hypoxia-inducible factor-3: Its structure, gene regulation and biological function (Review).Mol Med Rep. 2015 Aug;12(2):2411-6. doi: 10.3892/mmr.2015.3689. Epub 2015 Apr 27. Mol Med Rep. 2015. PMID: 25936862 Review.
-
Turn me on: regulating HIF transcriptional activity.Cell Death Differ. 2008 Apr;15(4):642-9. doi: 10.1038/sj.cdd.4402315. Epub 2008 Jan 18. Cell Death Differ. 2008. PMID: 18202699 Review.
Cited by
-
Hypoxia-induced transcription factor signaling is essential for larval growth of the mosquito Aedes aegypti.Proc Natl Acad Sci U S A. 2018 Jan 16;115(3):457-465. doi: 10.1073/pnas.1719063115. Epub 2018 Jan 3. Proc Natl Acad Sci U S A. 2018. PMID: 29298915 Free PMC article.
-
Reduced Cardiac Calcineurin Expression Mimics Long-Term Hypoxia-Induced Heart Defects in Drosophila.Circ Cardiovasc Genet. 2017 Oct;10(5):e001706. doi: 10.1161/CIRCGENETICS.117.001706. Circ Cardiovasc Genet. 2017. PMID: 28986453 Free PMC article.
-
Bottlenecks caused by software gaps in miRNA and RNAi research.Pharm Res. 2012 Jul;29(7):1717-21. doi: 10.1007/s11095-012-0712-x. Epub 2012 Feb 24. Pharm Res. 2012. PMID: 22362409
-
Musashi mediates translational repression of the Drosophila hypoxia inducible factor.Nucleic Acids Res. 2016 Sep 19;44(16):7555-67. doi: 10.1093/nar/gkw372. Epub 2016 May 3. Nucleic Acids Res. 2016. PMID: 27141964 Free PMC article.
-
Genome-scale RNAi on living-cell microarrays identifies novel regulators of Drosophila melanogaster TORC1-S6K pathway signaling.Genome Res. 2011 Mar;21(3):433-46. doi: 10.1101/gr.111492.110. Epub 2011 Jan 14. Genome Res. 2011. PMID: 21239477 Free PMC article.
References
-
- Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. - PubMed
-
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. - PubMed
-
- Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem. 2002;277:26351–26355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

