Identification of conserved regions and residues within Hedgehog acyltransferase critical for palmitoylation of Sonic Hedgehog
- PMID: 20585641
- PMCID: PMC2890405
- DOI: 10.1371/journal.pone.0011195
Identification of conserved regions and residues within Hedgehog acyltransferase critical for palmitoylation of Sonic Hedgehog
Abstract
Background: Sonic hedgehog (Shh) is a palmitoylated protein that plays key roles in mammalian development and human cancers. Palmitoylation of Shh is required for effective long and short range Shh-mediated signaling. Attachment of palmitate to Shh is catalyzed by Hedgehog acyltransferase (Hhat), a member of the membrane bound O-acyl transferase (MBOAT) family of multipass membrane proteins. The extremely hydrophobic composition of MBOAT proteins has limited their biochemical characterization. Except for mutagenesis of two conserved residues, there has been no structure-function analysis of Hhat, and the regions of the protein required for Shh palmitoylation are unknown.
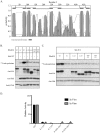
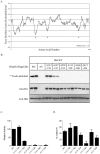
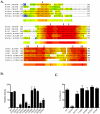
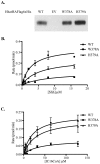
Methodology/principal findings: Here we undertake a systematic approach to identify residues within Hhat that are required for protein stability and/or enzymatic activity. We also identify a second, novel MBOAT homology region (residues 196-234) that is required for Hhat activity. In total, ten deletion mutants and eleven point mutants were generated and analyzed. Truncations at the N- and C-termini of Hhat yielded inactive proteins with reduced stability. Four Hhat mutants with deletions within predicted loop regions and five point mutants retained stability but lost palmitoylation activity. We purified two point mutants, W378A and H379A, with defective Hhat activity. Kinetic analyses revealed alterations in apparent K(m) and V(max) for Shh and/or palmitoyl CoA, changes that likely explain the catalytic defects observed for these mutants.
Conclusions/significance: This study has pinpointed specific regions and multiple residues that regulate Hhat stability and catalysis. Our findings should be applicable to other MBOAT proteins that mediate lipid modification of Wnt proteins and ghrelin, and should serve as a model for understanding how secreted morphogens are modified by palmitoyl acyltransferases.
Conflict of interest statement
Figures

Similar articles
-
Palmitoylation of Hedgehog proteins.Vitam Horm. 2012;88:229-52. doi: 10.1016/B978-0-12-394622-5.00010-9. Vitam Horm. 2012. PMID: 22391306 Free PMC article. Review.
-
Palmitoylation of Hedgehog proteins by Hedgehog acyltransferase: roles in signalling and disease.Open Biol. 2021 Mar;11(3):200414. doi: 10.1098/rsob.200414. Epub 2021 Mar 3. Open Biol. 2021. PMID: 33653085 Free PMC article. Review.
-
Membrane topology of hedgehog acyltransferase.J Biol Chem. 2015 Jan 23;290(4):2235-43. doi: 10.1074/jbc.M114.625764. Epub 2014 Dec 8. J Biol Chem. 2015. PMID: 25488661 Free PMC article.
-
Identification of N-terminal residues of Sonic Hedgehog important for palmitoylation by Hedgehog acyltransferase.J Biol Chem. 2012 Dec 14;287(51):42881-9. doi: 10.1074/jbc.M112.426833. Epub 2012 Oct 30. J Biol Chem. 2012. PMID: 23112049 Free PMC article.
-
Hedgehog acyltransferase catalyzes a random sequential reaction and utilizes multiple fatty acyl-CoA substrates.J Biol Chem. 2022 Oct;298(10):102422. doi: 10.1016/j.jbc.2022.102422. Epub 2022 Aug 24. J Biol Chem. 2022. PMID: 36030053 Free PMC article.
Cited by
-
Ghrelin O-acyltransferase assays and inhibition.Methods Enzymol. 2012;514:205-28. doi: 10.1016/B978-0-12-381272-8.00013-1. Methods Enzymol. 2012. PMID: 22975055 Free PMC article. Review.
-
Fatty acylation of Wnt proteins.Nat Chem Biol. 2016 Feb;12(2):60-9. doi: 10.1038/nchembio.2005. Nat Chem Biol. 2016. PMID: 26784846
-
Topological analysis of Hedgehog acyltransferase, a multipalmitoylated transmembrane protein.J Biol Chem. 2015 Feb 6;290(6):3293-307. doi: 10.1074/jbc.M114.614578. Epub 2014 Dec 12. J Biol Chem. 2015. PMID: 25505265 Free PMC article.
-
Palmitoylation of Hedgehog proteins.Vitam Horm. 2012;88:229-52. doi: 10.1016/B978-0-12-394622-5.00010-9. Vitam Horm. 2012. PMID: 22391306 Free PMC article. Review.
-
Determination of the membrane topology of PORCN, an O-acyl transferase that modifies Wnt signalling proteins.Open Biol. 2021 Jun;11(6):200400. doi: 10.1098/rsob.200400. Epub 2021 Jun 30. Open Biol. 2021. PMID: 34186010 Free PMC article.
References
-
- Ho KS, Scott MP. Sonic hedgehog in the nervous system: functions, modifications and mechanisms. Curr Opin Neurobiol. 2002;12:57–63. - PubMed
-
- Fuccillo M, Joyner AL, Fishell G. Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:772–783. - PubMed
-
- McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. - PubMed
-
- Lee JD, Kraus P, Gaiano N, Nery S, Kohtz J, et al. An acylatable residue of Hedgehog is differentially required in Drosophila and mouse limb development. Dev Biol. 2001;233:122–136. - PubMed
-
- Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

