Classification of dengue fever patients based on gene expression data using support vector machines
- PMID: 20585645
- PMCID: PMC2890409
- DOI: 10.1371/journal.pone.0011267
Classification of dengue fever patients based on gene expression data using support vector machines
Abstract
Background: Symptomatic infection by dengue virus (DENV) can range from dengue fever (DF) to dengue haemorrhagic fever (DHF), however, the determinants of DF or DHF progression are not completely understood. It is hypothesised that host innate immune response factors are involved in modulating the disease outcome and the expression levels of genes involved in this response could be used as early prognostic markers for disease severity.
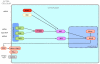
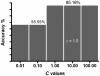
Methodology/principal findings: mRNA expression levels of genes involved in DENV innate immune responses were measured using quantitative real time PCR (qPCR). Here, we present a novel application of the support vector machines (SVM) algorithm to analyze the expression pattern of 12 genes in peripheral blood mononuclear cells (PBMCs) of 28 dengue patients (13 DHF and 15 DF) during acute viral infection. The SVM model was trained using gene expression data of these genes and achieved the highest accuracy of approximately 85% with leave-one-out cross-validation. Through selective removal of gene expression data from the SVM model, we have identified seven genes (MYD88, TLR7, TLR3, MDA5, IRF3, IFN-alpha and CLEC5A) that may be central in differentiating DF patients from DHF, with MYD88 and TLR7 observed to be the most important. Though the individual removal of expression data of five other genes had no impact on the overall accuracy, a significant combined role was observed when the SVM model of the two main genes (MYD88 and TLR7) was re-trained to include the five genes, increasing the overall accuracy to approximately 96%.
Conclusions/significance: Here, we present a novel use of the SVM algorithm to classify DF and DHF patients, as well as to elucidate the significance of the various genes involved. It was observed that seven genes are critical in classifying DF and DHF patients: TLR3, MDA5, IRF3, IFN-alpha, CLEC5A, and the two most important MYD88 and TLR7. While these preliminary results are promising, further experimental investigation is necessary to validate their specific roles in dengue disease.
Conflict of interest statement
Figures

Similar articles
-
Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever but not dengue hemorrhagic fever.J Infect Dis. 2008 May 15;197(10):1459-67. doi: 10.1086/587699. J Infect Dis. 2008. PMID: 18444802
-
Innate Lymphoid Cells Activation and Transcriptomic Changes in Response to Human Dengue Infection.Front Immunol. 2021 May 17;12:599805. doi: 10.3389/fimmu.2021.599805. eCollection 2021. Front Immunol. 2021. PMID: 34079535 Free PMC article. Clinical Trial.
-
Sequential waves of gene expression in patients with clinically defined dengue illnesses reveal subtle disease phases and predict disease severity.PLoS Negl Trop Dis. 2013 Jul 11;7(7):e2298. doi: 10.1371/journal.pntd.0002298. Print 2013. PLoS Negl Trop Dis. 2013. PMID: 23875036 Free PMC article.
-
Dengue and dengue haemorrhagic fever: implications of host genetics.FEMS Immunol Med Microbiol. 2006 Jul;47(2):155-66. doi: 10.1111/j.1574-695X.2006.00058.x. FEMS Immunol Med Microbiol. 2006. PMID: 16831202 Review.
-
Evolution of dengue in Sri Lanka-changes in the virus, vector, and climate.Int J Infect Dis. 2014 Feb;19:6-12. doi: 10.1016/j.ijid.2013.10.012. Epub 2013 Dec 11. Int J Infect Dis. 2014. PMID: 24334026 Review.
Cited by
-
Dengue and soluble mediators of the innate immune system.Trop Med Health. 2011 Dec;39(4 Suppl):53-62. doi: 10.2149/tmh.2011-S06. Epub 2011 Sep 13. Trop Med Health. 2011. PMID: 22500137 Free PMC article.
-
Potential biomarkers for the clinical prognosis of severe dengue.Mem Inst Oswaldo Cruz. 2013 Sep;108(6):755-62. doi: 10.1590/0074-0276108062013012. Mem Inst Oswaldo Cruz. 2013. PMID: 24037198 Free PMC article.
-
Differential expression of microRNA, miR-150 and enhancer of zeste homolog 2 (EZH2) in peripheral blood cells as early prognostic markers of severe forms of dengue.J Biomed Sci. 2020 Jan 18;27(1):25. doi: 10.1186/s12929-020-0620-z. J Biomed Sci. 2020. PMID: 31954402 Free PMC article.
-
Expression of Nitric Oxide Synthase and Nitric Oxide Levels in Peripheral Blood Cells and Oxidized Low-Density Lipoprotein Levels in Saliva as Early Markers of Severe Dengue.Biomed Res Int. 2021 Feb 9;2021:6650596. doi: 10.1155/2021/6650596. eCollection 2021. Biomed Res Int. 2021. PMID: 33628800 Free PMC article.
-
Predictive Models for the Medical Diagnosis of Dengue: A Case Study in Paraguay.Comput Math Methods Med. 2019 Jul 29;2019:7307803. doi: 10.1155/2019/7307803. eCollection 2019. Comput Math Methods Med. 2019. PMID: 31485259 Free PMC article.
References
-
- Holmes EC, Burch SS. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000;8:74–77. - PubMed
-
- Coffey LL, Mertens E, Brehin AC, Fernandez-Garcia MD, Amara A, et al. Human genetic determinants of dengue virus susceptibility. Microbes Infect. 2009;11:143–156. - PubMed
-
- Halstead SB. Dengue. Lancet. 2007;370:1644–1652. - PubMed
-
- WHO. 1997. Haemorrhagic Fever: Diagnosis, Treatment, Prevention and Control, second ed.
-
- Ubol S, Masrinoul P, Chaijaruwanich J, Kalayanarooj S, Charoensirisuthikul T, et al. Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever but not dengue hemorrhagic fever. J Infect Dis. 2008;197:1459–1467. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

