Medicinal chemistry of ATP synthase: a potential drug target of dietary polyphenols and amphibian antimicrobial peptides
- PMID: 20586714
- PMCID: PMC4734657
- DOI: 10.2174/092986710791859270
Medicinal chemistry of ATP synthase: a potential drug target of dietary polyphenols and amphibian antimicrobial peptides
Abstract
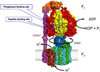
In this review we discuss the inhibitory effects of dietary polyphenols and amphibian antimicrobial/antitumor peptides on ATP synthase. In the beginning general structural features highlighting catalytic and motor functions of ATP synthase will be described. Some details on the presence of ATP synthase on the surface of several animal cell types, where it is associated with multiple cellular processes making it an interesting drug target with respect to dietary polyphenols and amphibian antimicrobial peptides will also be reviewed. ATP synthase is known to have distinct polyphenol and peptide binding sites at the interface of α/β subunits. Molecular interaction of polyphenols and peptides with ATP synthase at their respective binding sites will be discussed. Binding and inhibition of other proteins or enzymes will also be covered so as to understand the therapeutic roles of both types of molecules. Lastly, the effects of polyphenols and peptides on the inhibition of Escherichia coli cell growth through their action on ATP synthase will also be presented.
Figures





Similar articles
-
A Therapeutic Connection between Dietary Phytochemicals and ATP Synthase.Curr Med Chem. 2017 Nov 20;24(35):3894-3906. doi: 10.2174/0929867324666170823125330. Curr Med Chem. 2017. PMID: 28831918 Free PMC article. Review.
-
ATP synthase: a molecular therapeutic drug target for antimicrobial and antitumor peptides.Curr Med Chem. 2013;20(15):1956-73. doi: 10.2174/0929867311320150003. Curr Med Chem. 2013. PMID: 23432591 Free PMC article. Review.
-
Inhibition of Escherichia coli ATP synthase by amphibian antimicrobial peptides.Int J Biol Macromol. 2010 Apr 1;46(3):367-74. doi: 10.1016/j.ijbiomac.2010.01.015. Epub 2010 Jan 25. Int J Biol Macromol. 2010. PMID: 20100509 Free PMC article.
-
Reversible optical control of F1 Fo -ATP synthase using photoswitchable inhibitors.FEBS Lett. 2018 Feb;592(3):343-355. doi: 10.1002/1873-3468.12958. Epub 2018 Feb 1. FEBS Lett. 2018. PMID: 29292505 Free PMC article.
-
Thiol oxidation of mitochondrial F0-c subunits: a way to switch off antimicrobial drug targets of the mitochondrial ATP synthase.Med Hypotheses. 2014 Aug;83(2):160-5. doi: 10.1016/j.mehy.2014.05.004. Epub 2014 May 17. Med Hypotheses. 2014. PMID: 24932580
Cited by
-
Affinity-based target deconvolution of safranal.Daru. 2013 Mar 20;21(1):25. doi: 10.1186/2008-2231-21-25. Daru. 2013. PMID: 23514587 Free PMC article.
-
Interactions with Microbial Proteins Driving the Antibacterial Activity of Flavonoids.Pharmaceutics. 2021 May 5;13(5):660. doi: 10.3390/pharmaceutics13050660. Pharmaceutics. 2021. PMID: 34062983 Free PMC article. Review.
-
Autophagy and the peroxisome proliferator-activated receptor signaling pathway: A molecular ballet in lipid metabolism and homeostasis.Mol Cell Biochem. 2025 Jun;480(6):3477-3499. doi: 10.1007/s11010-025-05207-0. Epub 2025 Feb 1. Mol Cell Biochem. 2025. PMID: 39891864 Review.
-
A Therapeutic Connection between Dietary Phytochemicals and ATP Synthase.Curr Med Chem. 2017 Nov 20;24(35):3894-3906. doi: 10.2174/0929867324666170823125330. Curr Med Chem. 2017. PMID: 28831918 Free PMC article. Review.
-
Thymoquinone Inhibits Escherichia coli ATP Synthase and Cell Growth.PLoS One. 2015 May 21;10(5):e0127802. doi: 10.1371/journal.pone.0127802. eCollection 2015. PLoS One. 2015. PMID: 25996607 Free PMC article.
References
-
- Senior AE, Nadanaciva S, Weber J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta. 2002;1553:188–211. - PubMed
-
- Boyer PD. The ATP synthase-a splendid molecular machine. Annu. Rev. Biochem. 1997;66:717–749. - PubMed
-
- Van Walraven HS, Strotmann H, Schwarz O, Rumberg B. The H+/ATP coupling ratio of the ATP synthase from thiol-modulated chloroplasts and two cyanobacterial strains is four. FEBS Lett. 1996;379:309–313. - PubMed
-
- Yoshida M, Muneyuki E, Hisabori T. ATP synthase--a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001;2:669–677. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

