Clinical proteomics of myeloid leukemia
- PMID: 20587003
- PMCID: PMC2905101
- DOI: 10.1186/gm162
Clinical proteomics of myeloid leukemia
Abstract
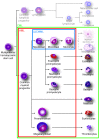
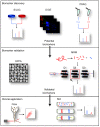
Myeloid leukemias are a heterogeneous group of diseases originating from bone marrow myeloid progenitor cells. Patients with myeloid leukemias can achieve long-term survival through targeted therapy, cure after intensive chemotherapy or short-term survival because of highly chemoresistant disease. Therefore, despite the development of advanced molecular diagnostics, there is an unmet need for efficient therapy that reflects the advanced diagnostics. Although the molecular design of therapeutic agents is aimed at interacting with specific proteins identified through molecular diagnostics, the majority of therapeutic agents act on multiple protein targets. Ongoing studies on the leukemic cell proteome will probably identify a large number of new biomarkers, and the prediction of response to therapy through these markers is an interesting avenue for future personalized medicine. Mass spectrometric protein detection is a fundamental technique in clinical proteomics, and selected tools are presented, including stable isotope labeling with amino acids in cell culture (SILAC), isobaric tags for relative and absolute quantification (iTRAQ) and multiple reaction monitoring (MRM), as well as single cell determination. We suggest that protein analysis will play not only a supplementary, but also a prominent role in future molecular diagnostics, and we outline how accurate knowledge of the molecular therapeutic targets can be used to monitor therapy response.
Figures
Similar articles
-
Integrated Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and Isobaric Tags for Relative and Absolute Quantitation (iTRAQ) Quantitative Proteomic Analysis Identifies Galectin-1 as a Potential Biomarker for Predicting Sorafenib Resistance in Liver Cancer.Mol Cell Proteomics. 2015 Jun;14(6):1527-45. doi: 10.1074/mcp.M114.046417. Epub 2015 Apr 7. Mol Cell Proteomics. 2015. PMID: 25850433 Free PMC article. Clinical Trial.
-
Biomarker verification using selected reaction monitoring and shotgun proteomics.Methods Mol Biol. 2014;1156:295-306. doi: 10.1007/978-1-4939-0685-7_20. Methods Mol Biol. 2014. PMID: 24791997
-
Proteomics-based strategy to identify biomarkers and pharmacological targets in leukemias with t(4;11) translocations.J Proteome Res. 2006 Oct;5(10):2743-53. doi: 10.1021/pr060235v. J Proteome Res. 2006. PMID: 17022645
-
Proteomics approaches for myeloid leukemia drug discovery.Expert Opin Drug Discov. 2012 Dec;7(12):1165-75. doi: 10.1517/17460441.2012.724055. Epub 2012 Sep 13. Expert Opin Drug Discov. 2012. PMID: 22971110 Review.
-
An insight into iTRAQ: where do we stand now?Anal Bioanal Chem. 2012 Sep;404(4):1011-27. doi: 10.1007/s00216-012-5918-6. Epub 2012 Mar 27. Anal Bioanal Chem. 2012. PMID: 22451173 Review.
Cited by
-
Discovery and development of the Polo-like kinase inhibitor volasertib in cancer therapy.Leukemia. 2015 Jan;29(1):11-9. doi: 10.1038/leu.2014.222. Epub 2014 Jul 16. Leukemia. 2015. PMID: 25027517 Free PMC article. Review.
-
Proteomics-based discovery of biomarkers for paediatric acute lymphoblastic leukaemia: challenges and opportunities.J Cell Mol Med. 2014 Jul;18(7):1239-46. doi: 10.1111/jcmm.12319. Epub 2014 Jun 9. J Cell Mol Med. 2014. PMID: 24912534 Free PMC article. Review.
-
Highly multiplexed proteomic assessment of human bone marrow in acute myeloid leukemia.Blood Adv. 2020 Jan 28;4(2):367-379. doi: 10.1182/bloodadvances.2019001124. Blood Adv. 2020. PMID: 31985806 Free PMC article.
-
Specific cellular signal-transduction responses to in vivo combination therapy with ATRA, valproic acid and theophylline in acute myeloid leukemia.Blood Cancer J. 2011 Feb;1(2):e4. doi: 10.1038/bcj.2011.2. Epub 2011 Feb 11. Blood Cancer J. 2011. PMID: 22829110 Free PMC article.
-
Antibody-based detection of protein phosphorylation status to track the efficacy of novel therapies using nanogram protein quantities from stem cells and cell lines.Nat Protoc. 2015 Jan;10(1):149-68. doi: 10.1038/nprot.2015.007. Epub 2014 Dec 18. Nat Protoc. 2015. PMID: 25521791
References
-
- Hochhaus A, O'Brien SG, Guilhot F, Druker BJ, Branford S, Foroni L, Goldman JM, Muller MC, Radich JP, Rudoltz M, Mone M, Gathmann I, Hughes TP, Larson RA. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–1061. doi: 10.1038/leu.2009.38. - DOI - PubMed
-
- Rix U, Hantschel O, Durnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, Kocher T, Superti-Furga G. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. - DOI - PubMed
LinkOut - more resources
Full Text Sources

