Comparing large-scale computational approaches to epidemic modeling: agent-based versus structured metapopulation models
- PMID: 20587041
- PMCID: PMC2914769
- DOI: 10.1186/1471-2334-10-190
Comparing large-scale computational approaches to epidemic modeling: agent-based versus structured metapopulation models
Abstract
Background: In recent years large-scale computational models for the realistic simulation of epidemic outbreaks have been used with increased frequency. Methodologies adapt to the scale of interest and range from very detailed agent-based models to spatially-structured metapopulation models. One major issue thus concerns to what extent the geotemporal spreading pattern found by different modeling approaches may differ and depend on the different approximations and assumptions used.
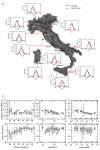
Methods: We provide for the first time a side-by-side comparison of the results obtained with a stochastic agent-based model and a structured metapopulation stochastic model for the progression of a baseline pandemic event in Italy, a large and geographically heterogeneous European country. The agent-based model is based on the explicit representation of the Italian population through highly detailed data on the socio-demographic structure. The metapopulation simulations use the GLobal Epidemic and Mobility (GLEaM) model, based on high-resolution census data worldwide, and integrating airline travel flow data with short-range human mobility patterns at the global scale. The model also considers age structure data for Italy. GLEaM and the agent-based models are synchronized in their initial conditions by using the same disease parameterization, and by defining the same importation of infected cases from international travels.
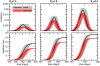
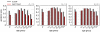
Results: The results obtained show that both models provide epidemic patterns that are in very good agreement at the granularity levels accessible by both approaches, with differences in peak timing on the order of a few days. The relative difference of the epidemic size depends on the basic reproductive ratio, R0, and on the fact that the metapopulation model consistently yields a larger incidence than the agent-based model, as expected due to the differences in the structure in the intra-population contact pattern of the approaches. The age breakdown analysis shows that similar attack rates are obtained for the younger age classes.
Conclusions: The good agreement between the two modeling approaches is very important for defining the tradeoff between data availability and the information provided by the models. The results we present define the possibility of hybrid models combining the agent-based and the metapopulation approaches according to the available data and computational resources.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

