Evolution of seventh cholera pandemic and origin of 1991 epidemic, Latin America
- PMID: 20587187
- PMCID: PMC3321917
- DOI: 10.3201/eid1607.100131
Evolution of seventh cholera pandemic and origin of 1991 epidemic, Latin America
Abstract
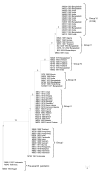
Thirty single-nucleotide polymorphisms were used to track the spread of the seventh pandemic caused by Vibrio cholerae. Isolates from the 1991 epidemic in Latin America shared a profile with 1970s isolates from Africa, suggesting a possible origin in Africa. Data also showed that the observed genotypes spread easily and widely.
Figures
References
-
- Reeves PR, Lan R. Cholera in the 1990s. Br Med Bull. 1998;54:611–23. - PubMed
-
- Seas C, Miranda J, Gil AI, Leon-Barua R, Patz J, Huq A, et al. New insights on the emergence of cholera in Latin America during 1991: the Peruvian experience. Am J Trop Med Hyg. 2000;62:513–7. - PubMed
-
- Singh DV, Matte MH, Matte GR, Jiang S, Sabeena F, Shukla BN, et al. Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl Environ Microbiol. 2001;67:910–21. 10.1128/AEM.67.2.910-921.2001 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
