p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals
- PMID: 20587414
- PMCID: PMC2937881
- DOI: 10.1074/jbc.M110.149393
p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals
Abstract
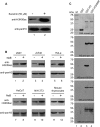
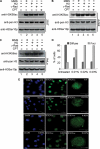
The packaging of newly replicated and repaired DNA into chromatin is crucial for the maintenance of genomic integrity. Acetylation of histone H3 core domain lysine 56 (H3K56ac) has been shown to play a crucial role in compaction of DNA into chromatin following replication and repair in Saccharomyces cerevisiae. However, the occurrence and function of such acetylation has not been reported in mammals. Here we show that H3K56 is acetylated and that this modification is regulated in a cell cycle-dependent manner in mammalian cells. We also demonstrate that the histone acetyltransferase p300 acetylates H3K56 in vitro and in vivo, whereas hSIRT2 and hSIRT3 deacetylate H3K56ac in vivo. Further we show that following DNA damage H3K56 acetylation levels increased, and acetylated H3K56, which is localized at the sites of DNA repair. It also colocalized with other proteins involved in DNA damage signaling pathways such as phospho-ATM, CHK2, and p53. Interestingly, analysis of occurrence of H3K56 acetylation using ChIP-on-chip revealed its genome-wide spread, affecting genes involved in several pathways that are implicated in tumorigenesis such as cell cycle, DNA damage response, DNA repair, and apoptosis.
Figures





References
-
- Ehrenhofer-Murray A. E. (2004) Eur. J. Biochem. 271, 2335–2349 - PubMed
-
- Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 - PubMed
-
- Peterson C. L., Laniel M. A. (2004). Curr. Biol. 14, R546–R551 - PubMed
-
- Bird A. W., Yu D. Y., Pray-Grant M. G., Qiu Q., Harmon K. E., Megee P. C., Grant P. A., Smith M. M., Christman M. F. (2002) Nature 419, 411–415 - PubMed
-
- Kouzarides T. (2007) Cell 128, 693–705 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

