When simple sequence comparison fails: the cryptic case of the shared domains of the bacterial replication initiation proteins DnaB and DnaD
- PMID: 20587500
- PMCID: PMC2978336
- DOI: 10.1093/nar/gkq465
When simple sequence comparison fails: the cryptic case of the shared domains of the bacterial replication initiation proteins DnaB and DnaD
Abstract
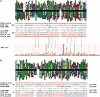
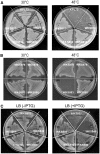
DnaD and DnaB are essential DNA-replication-initiation proteins in low-G+C content Gram-positive bacteria. Here we use sensitive Hidden Markov Model-based techniques to show that the DnaB and DnaD proteins share a common structure that is evident across all their structural domains, termed DDBH1 and DDBH2 (DnaD DnaB Homology 1 and 2). Despite strong sequence divergence, many of the DNA-binding and oligomerization properties of these domains have been conserved. Although eluding simple sequence comparisons, the DDBH2 domains share the only strong sequence motif; an extremely highly conserved YxxxIxxxW sequence that contributes to DNA binding. Sequence alignments of DnaD alone fail to identify another key part of the DNA-binding module, since it includes a poorly conserved sequence, a solvent-exposed and somewhat unstable helix and a mobile segment. We show by NMR, in vitro mutagenesis and in vivo complementation experiments that the DNA-binding module of Bacillus subtilis DnaD comprises the YxxxIxxxW motif, the unstable helix and a portion of the mobile region, the latter two being essential for viability. These structural insights lead us to a re-evaluation of the oligomerization and DNA-binding properties of the DnaD and DnaB proteins.
Figures




Similar articles
-
DNA replication initiation in Bacillus subtilis: structural and functional characterization of the essential DnaA-DnaD interaction.Nucleic Acids Res. 2019 Feb 28;47(4):2101-2112. doi: 10.1093/nar/gky1220. Nucleic Acids Res. 2019. PMID: 30534966 Free PMC article.
-
Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication.Mol Microbiol. 2005 Feb;55(4):1138-50. doi: 10.1111/j.1365-2958.2004.04451.x. Mol Microbiol. 2005. PMID: 15686560
-
Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis.Mol Microbiol. 2004 Jun;52(6):1757-67. doi: 10.1111/j.1365-2958.2004.04091.x. Mol Microbiol. 2004. PMID: 15186423
-
New types of conserved sequence domains in DNA-binding regions of homing endonucleases.Trends Biochem Sci. 2003 Sep;28(9):473-7. doi: 10.1016/S0968-0004(03)00170-1. Trends Biochem Sci. 2003. PMID: 13678957 Review.
-
Structural Insight Into the Function of DnaB Helicase in Bacterial DNA Replication.Proteins. 2025 Feb;93(2):420-429. doi: 10.1002/prot.26746. Epub 2024 Sep 4. Proteins. 2025. PMID: 39230358 Review.
Cited by
-
Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target.Curr Drug Targets. 2012 Mar;13(3):352-72. doi: 10.2174/138945012799424598. Curr Drug Targets. 2012. PMID: 22206257 Free PMC article. Review.
-
The primosomal protein DnaD inhibits cooperative DNA binding by the replication initiator DnaA in Bacillus subtilis.J Bacteriol. 2012 Sep;194(18):5110-7. doi: 10.1128/JB.00958-12. Epub 2012 Jul 20. J Bacteriol. 2012. PMID: 22821970 Free PMC article.
-
Control of Initiation of DNA Replication in Bacillus subtilis and Escherichia coli.Genes (Basel). 2017 Jan 10;8(1):22. doi: 10.3390/genes8010022. Genes (Basel). 2017. PMID: 28075389 Free PMC article. Review.
-
Staphylococcus aureus single-stranded DNA-binding protein SsbA can bind but cannot stimulate PriA helicase.PLoS One. 2017 Jul 27;12(7):e0182060. doi: 10.1371/journal.pone.0182060. eCollection 2017. PLoS One. 2017. PMID: 28750050 Free PMC article.
-
Loading mechanisms of ring helicases at replication origins.Mol Microbiol. 2012 Apr;84(1):6-16. doi: 10.1111/j.1365-2958.2012.08012.x. Epub 2012 Mar 15. Mol Microbiol. 2012. PMID: 22417087 Free PMC article. Review.
References
-
- Marsin S, McGovern S, Ehrlic HSD, Bruand C, Polard P. Early steps of Bacillus subtilis primosome assembly. J. Biol. Chem. 2001;276:45818–45825. - PubMed
-
- Bruand C, Velten M, McGovern S, Marsin S, Serena C, Ehrlich SD, Polard P. Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication. Mol. Microbiol. 2005;55:1138–1150. - PubMed
-
- Mott ML, Berger M. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007;5:343–354. - PubMed
-
- Zakrzewska-Czerwinska J, Jakimowicz D, Zawilak-Pawlik A, Messer W. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol. Rev. 2007;31:378–387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

