DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis
- PMID: 20587527
- PMCID: PMC2905468
- DOI: 10.1158/0008-5472.CAN-10-0847
DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis
Abstract
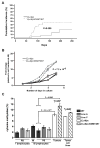
Epigenetic changes are among the most common alterations observed in cancer cells, yet the mechanism by which cancer cells acquire and maintain abnormal DNA methylation patterns is not understood. Cancer cells have an altered distribution of DNA methylation and express aberrant DNA methyltransferase 3B transcripts, which encode truncated proteins, some of which lack the COOH-terminal catalytic domain. To test if a truncated DNMT3B isoform disrupts DNA methylation in vivo, we constructed two lines of transgenic mice expressing DNMT3B7, a truncated DNMT3B isoform commonly found in cancer cells. DNMT3B7 transgenic mice exhibit altered embryonic development, including lymphopenia, craniofacial abnormalities, and cardiac defects, similar to Dnmt3b-deficient animals, but rarely develop cancer. However, when DNMT3B7 transgenic mice are bred with Emicro-Myc transgenic mice, which model aggressive B-cell lymphoma, DNMT3B7 expression increases the frequency of mediastinal lymphomas in Emicro-Myc animals. Emicro-Myc/DNMT3B7 mediastinal lymphomas have more chromosomal rearrangements, increased global DNA methylation levels, and more locus-specific perturbations in DNA methylation patterns compared with Emicro-Myc lymphomas. These data represent the first in vivo modeling of cancer-associated DNA methylation changes and suggest that truncated DNMT3B isoforms contribute to the redistribution of DNA methylation characterizing virtually every human tumor.
(c)2010 AACR.
Figures




References
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. - PubMed
-
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

