Comprehensive analysis of the palindromic motif TCTCGCGAGA: a regulatory element of the HNRNPK promoter
- PMID: 20587588
- PMCID: PMC2920758
- DOI: 10.1093/dnares/dsq016
Comprehensive analysis of the palindromic motif TCTCGCGAGA: a regulatory element of the HNRNPK promoter
Abstract
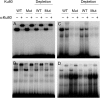
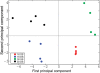
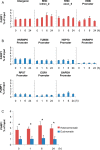
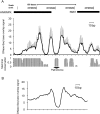
Definitive identification of promoters, their cis-regulatory motifs, and their trans-acting proteins requires experimental analysis. To define the HNRNPK promoter and its cognate DNA-protein interactions, we performed a comprehensive study combining experimental approaches, including luciferase reporter gene assays, chromatin immunoprecipitations (ChIP), electrophoretic mobility shift assays (EMSA), and mass spectrometry (MS). We discovered that out of the four potential HNRNPK promoters tested, the one containing the palindromic motif TCTCGCGAGA exhibited the highest activity in a reporter system assay. Although further EMSA and MS analyses, performed to uncover the identity of the palindrome-binding transcription factor, did identify a complex of DNA-binding proteins, neither method unambiguously identified the pertinent direct trans-acting protein(s). ChIP revealed similar chromatin states at the promoters with the palindromic motif and at housekeeping gene promoters. A ChIP survey showed significantly higher recruitment of PARP1, a protein identified by MS as ubiquitously attached to DNA probes, within heterochromatin sites. Computational analyses indicated that this palindrome displays features that mark nucleosome boundaries, causing the surrounding DNA landscape to be constitutively open. Our strategy of diverse approaches facilitated the direct characterization of various molecular properties of HNRNPK promoter bearing the palindromic motif TCTCGCGAGA, despite the obstacles that accompany in vitro methods.
Figures



Similar articles
-
Cloning and characterization of the human SH3BP2 promoter.Biochem Biophys Res Commun. 2012 Aug 17;425(1):25-32. doi: 10.1016/j.bbrc.2012.07.043. Epub 2012 Jul 17. Biochem Biophys Res Commun. 2012. PMID: 22820184 Free PMC article.
-
Human sodium-iodide symporter (hNIS) gene expression is inhibited by a trans-active transcriptional repressor, NIS-repressor, containing PARP-1 in thyroid cancer cells.Endocr Relat Cancer. 2010 Apr 21;17(2):383-98. doi: 10.1677/ERC-09-0156. Print 2010 Jun. Endocr Relat Cancer. 2010. PMID: 20228127
-
Characterization of a palindromic enhancer element in the promoters of IL4, IL5, and IL13 cytokine genes.J Allergy Clin Immunol. 2003 Apr;111(4):826-32. doi: 10.1067/mai.2003.1377. J Allergy Clin Immunol. 2003. PMID: 12704365
-
Expression of SMARCB1 modulates steroid sensitivity in human lymphoblastoid cells: identification of a promoter SNP that alters PARP1 binding and SMARCB1 expression.Hum Mol Genet. 2007 Oct 1;16(19):2261-71. doi: 10.1093/hmg/ddm178. Epub 2007 Jul 5. Hum Mol Genet. 2007. PMID: 17616514
-
PARP goes transcription.Cell. 2003 Jun 13;113(6):677-83. doi: 10.1016/s0092-8674(03)00433-1. Cell. 2003. PMID: 12809599 Review.
Cited by
-
Global analysis of S-nitrosylation sites in the wild type (APP) transgenic mouse brain-clues for synaptic pathology.Mol Cell Proteomics. 2014 Sep;13(9):2288-305. doi: 10.1074/mcp.M113.036079. Epub 2014 Jun 3. Mol Cell Proteomics. 2014. PMID: 24895380 Free PMC article.
-
Hypoxic 3D in vitro culture models reveal distinct resistance processes to TKIs in renal cancer cells.Cell Biosci. 2017 Dec 16;7:71. doi: 10.1186/s13578-017-0197-8. eCollection 2017. Cell Biosci. 2017. PMID: 29270287 Free PMC article.
-
The interactome of multifunctional HAX1 protein suggests its role in the regulation of energy metabolism, de-aggregation, cytoskeleton organization and RNA-processing.Biosci Rep. 2020 Nov 27;40(11):BSR20203094. doi: 10.1042/BSR20203094. Biosci Rep. 2020. PMID: 33146709 Free PMC article.
-
Mass Spectrometry-Based Comprehensive Analysis of Pancreatic Cyst Fluids.Biomed Res Int. 2018 Nov 29;2018:7169595. doi: 10.1155/2018/7169595. eCollection 2018. Biomed Res Int. 2018. PMID: 30627566 Free PMC article. Clinical Trial.
-
Conserved 3' UTR stem-loop structure in L1 and Alu transposons in human genome: possible role in retrotransposition.BMC Genomics. 2016 Dec 3;17(1):992. doi: 10.1186/s12864-016-3344-4. BMC Genomics. 2016. PMID: 27914481 Free PMC article.
References
-
- Bomsztyk K., Denisenko O., Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–38. doi:10.1002/bies.20048. - DOI - PubMed
-
- Habelhah H., Shah K., Huang L., et al. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell biol. 2001;3:325–30. doi:10.1038/35060131. - DOI - PubMed
-
- Moumen A., Masterson P., O'Connor M.J., Jackson S.P. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–78. doi:10.1016/j.cell.2005.09.032. - DOI - PubMed
-
- Carpenter B., McKay M., Dundas S.R., Lawrie L.C., Telfer C., Murray G.I. Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br. J. Cancer. 2006;95:921–7. doi:10.1038/sj.bjc.6603349. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

