Genome-wide computational identification and manual annotation of human long noncoding RNA genes
- PMID: 20587619
- PMCID: PMC2905748
- DOI: 10.1261/rna.1951310
Genome-wide computational identification and manual annotation of human long noncoding RNA genes
Abstract
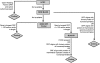
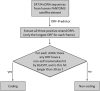
Experimental evidence suggests that half or more of the mammalian transcriptome consists of noncoding RNA. Noncoding RNAs are divided into short noncoding RNAs (including microRNAs) and long noncoding RNAs (lncRNAs). We defined complementary DNAs (cDNAs) lacking any positive-strand open reading frames (ORFs) longer than 30 amino acids, as well as cDNAs lacking any evidence of interspecies conservation of their longer-than-30-amino acid ORFs, as noncoding. We have identified 5446 lncRNA genes in the human genome from approximately 24,000 full-length cDNAs, using our new ORF-prediction pipeline. We combined them nonredundantly with lncRNAs from four published sources to derive 6736 lncRNA genes. In an effort to distinguish standalone and antisense lncRNA genes from database artifacts, we stratified our catalog of lncRNAs according to the distance between each lncRNA gene candidate and its nearest known protein-coding gene. We concurrently examined the protein-coding capacity of known genes overlapping with lncRNAs. Remarkably, 62% of known genes with "hypothetical protein" names actually lacked protein-coding capacity. This study has greatly expanded the known human lncRNA catalog, increased its accuracy through manual annotation of cDNA-to-genome alignments, and revealed that a large set of hypothetical-protein genes in GenBank lacks protein-coding capacity. In addition, we have developed, independently of existing NCBI tools, command-line programs with high-throughput ORF-finding and BLASTP-parsing functionality, suitable for future automated assessments of protein-coding capacity of novel transcripts.
Figures



References
-
- Carninci P, Hayashizaki Y 2007. Noncoding RNA transcription beyond annotated genes. Curr Opin Genet Dev 17: 139–144 - PubMed
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
