Sho1 and Msb2-related proteins regulate appressorium development in the smut fungus Ustilago maydis
- PMID: 20587773
- PMCID: PMC2910971
- DOI: 10.1105/tpc.109.073734
Sho1 and Msb2-related proteins regulate appressorium development in the smut fungus Ustilago maydis
Abstract
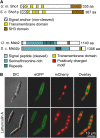
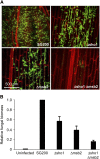
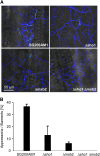
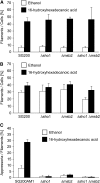
The dimorphic fungus Ustilago maydis switches from budding to hyphal growth on the plant surface. In response to hydrophobicity and hydroxy fatty acids, U. maydis develops infection structures called appressoria. Here, we report that, unlike in Saccharomyces cerevisiae and other fungi where Sho1 (synthetic high osmolarity sensitive) and Msb2 (multicopy suppressor of a budding defect) regulate stress responses and pseudohyphal growth, Sho1 and Msb2-like proteins play a key role during appressorium differentiation in U. maydis. Sho1 was identified through a two-hybrid screen as an interaction partner of the mitogen-activated protein (MAP) kinase Kpp6. Epistasis analysis revealed that sho1 and msb2 act upstream of the MAP kinases kpp2 and kpp6. Furthermore, Sho1 was shown to destabilize Kpp6 through direct interaction with the unique N-terminal domain in Kpp6, indicating a role of Sho1 in fine-tuning Kpp6 activity. Morphological differentiation in response to a hydrophobic surface was strongly attenuated in sho1 msb2 mutants, while hydroxy fatty acid-induced differentiation was unaffected. These data suggest that Sho1 and the transmembrane mucin Msb2 are involved in plant surface sensing in U. maydis.
Figures


References
-
- Aichinger C., Hansson K., Eichhorn H., Lessing F., Mannhaupt G., Mewes W., Kahmann R. (2003). Identification of plant-regulated genes in Ustilago maydis by enhancer-trapping mutagenesis. Mol. Genet. Genomics 270: 303–314 - PubMed
-
- Andrews D.L., Egan J.D., Mayorga M.E., Gold S.E. (2000). The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant Microbe Interact. 13: 781–786 - PubMed
-
- Banuett F., Herskowitz I. (1994). Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8: 1367–1378 - PubMed
-
- Bechinger C., Giebel K.F., Schnell M., Leiderer P., Deising H.B., Bastmeyer M. (1999). Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science 285: 1896–1899 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

