Mathematical modeling of endocytic actin patch kinetics in fission yeast: disassembly requires release of actin filament fragments
- PMID: 20587776
- PMCID: PMC2921120
- DOI: 10.1091/mbc.E10-06-0494
Mathematical modeling of endocytic actin patch kinetics in fission yeast: disassembly requires release of actin filament fragments
Abstract
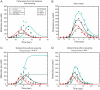
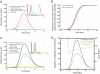
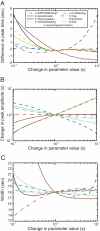
We used the dendritic nucleation hypothesis to formulate a mathematical model of the assembly and disassembly of actin filaments at sites of clathrin-mediated endocytosis in fission yeast. We used the wave of active WASp recruitment at the site of the patch formation to drive assembly reactions after activation of Arp2/3 complex. Capping terminated actin filament elongation. Aging of the filaments by ATP hydrolysis and gamma-phosphate dissociation allowed actin filament severing by cofilin. The model could simulate the assembly and disassembly of actin and other actin patch proteins using measured cytoplasmic concentrations of the proteins. However, to account quantitatively for the numbers of proteins measured over time in the accompanying article (Sirotkin et al., 2010, MBoC 21: 2792-2802), two reactions must be faster in cells than in vitro. Conditions inside the cell allow capping protein to bind to the barbed ends of actin filaments and Arp2/3 complex to bind to the sides of filaments faster than the purified proteins in vitro. Simulations also show that depolymerization from pointed ends cannot account for rapid loss of actin filaments from patches in 10 s. An alternative mechanism consistent with the data is that severing produces short fragments that diffuse away from the patch.
Figures


References
-
- Achard V., Martiel J. L., Michelot A., Guerin C., Reymann A. C., Blanchoin L., Boujemaa-Paterski R. A “primer”-based mechanism underlies branched actin filament network formation and motility. Curr. Biol. 2010;20:423–428. - PubMed
-
- Andrianantoandro E., Pollard T. D. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 2006;24:13–23. - PubMed
-
- Beltzner C.C., Pollard T. D. Pathway of actin filament branch formation by Arp2/3 complex. J. Biol. Chem. 2008;283:7135–7144. - PubMed
-
- Blanchoin L., Pollard T. D. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 1999;274:15538–15546. - PubMed
-
- Blanchoin L., Pollard T. D. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 2002;41:597–602. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

