Protein-protein interactions essentials: key concepts to building and analyzing interactome networks
- PMID: 20589078
- PMCID: PMC2891586
- DOI: 10.1371/journal.pcbi.1000807
Protein-protein interactions essentials: key concepts to building and analyzing interactome networks
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

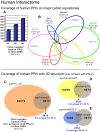
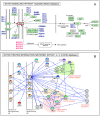
References
-
- Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum Mol Genet. 2005;14 Spec No. 2:R171–R181. - PubMed
-
- Blow N. Systems biology: Untangling the protein web. Nature. 2009;460:415–418. - PubMed
-
- Mackay JP, Sunde M, Lowry JA, Crossley M, Matthews JM. Protein interactions: is seeing believing? Trends Biochem Sci. 2007;32:530–531. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

