Use of linear mixed models for genetic evaluation of gestation length and birth weight allowing for heavy-tailed residual effects
- PMID: 20591149
- PMCID: PMC2909158
- DOI: 10.1186/1297-9686-42-26
Use of linear mixed models for genetic evaluation of gestation length and birth weight allowing for heavy-tailed residual effects
Abstract
Background: The distribution of residual effects in linear mixed models in animal breeding applications is typically assumed normal, which makes inferences vulnerable to outlier observations. In order to mute the impact of outliers, one option is to fit models with residuals having a heavy-tailed distribution. Here, a Student's-t model was considered for the distribution of the residuals with the degrees of freedom treated as unknown. Bayesian inference was used to investigate a bivariate Student's-t (BSt) model using Markov chain Monte Carlo methods in a simulation study and analysing field data for gestation length and birth weight permitted to study the practical implications of fitting heavy-tailed distributions for residuals in linear mixed models.
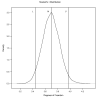
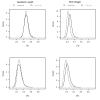
Methods: In the simulation study, bivariate residuals were generated using Student's-t distribution with 4 or 12 degrees of freedom, or a normal distribution. Sire models with bivariate Student's-t or normal residuals were fitted to each simulated dataset using a hierarchical Bayesian approach. For the field data, consisting of gestation length and birth weight records on 7,883 Italian Piemontese cattle, a sire-maternal grandsire model including fixed effects of sex-age of dam and uncorrelated random herd-year-season effects were fitted using a hierarchical Bayesian approach. Residuals were defined to follow bivariate normal or Student's-t distributions with unknown degrees of freedom.
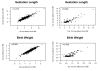
Results: Posterior mean estimates of degrees of freedom parameters seemed to be accurate and unbiased in the simulation study. Estimates of sire and herd variances were similar, if not identical, across fitted models. In the field data, there was strong support based on predictive log-likelihood values for the Student's-t error model. Most of the posterior density for degrees of freedom was below 4. Posterior means of direct and maternal heritabilities for birth weight were smaller in the Student's-t model than those in the normal model. Re-rankings of sires were observed between heavy-tailed and normal models.
Conclusions: Reliable estimates of degrees of freedom were obtained in all simulated heavy-tailed and normal datasets. The predictive log-likelihood was able to distinguish the correct model among the models fitted to heavy-tailed datasets. There was no disadvantage of fitting a heavy-tailed model when the true model was normal. Predictive log-likelihood values indicated that heavy-tailed models with low degrees of freedom values fitted gestation length and birth weight data better than a model with normally distributed residuals.Heavy-tailed and normal models resulted in different estimates of direct and maternal heritabilities, and different sire rankings. Heavy-tailed models may be more appropriate for reliable estimation of genetic parameters from field data.
Figures


References
-
- Roger WH, W TJ. Understanding some long-tailed distributions. Statistica Neerlandia. 1972;26:211 226.
-
- Lange KL, Little RJA, Taylor JMG. Robust Statistical Modeling Using the t Distribution. J Am Stat Assoc. 1989;84:881–896. doi: 10.2307/2290063. - DOI
-
- Stranden I, Gianola D. Mixed effects linear models with t-distributions for quantitative genetic analysis: a Bayesian approach. Genet Sel Evol. 1999;31:25–42. doi: 10.1186/1297-9686-31-1-25. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

