Automated quantitative live cell fluorescence microscopy
- PMID: 20591990
- PMCID: PMC2908775
- DOI: 10.1101/cshperspect.a000455
Automated quantitative live cell fluorescence microscopy
Abstract
Advances in microscopy automation and image analysis have given biologists the tools to attempt large scale systems-level experiments on biological systems using microscope image readout. Fluorescence microscopy has become a standard tool for assaying gene function in RNAi knockdown screens and protein localization studies in eukaryotic systems. Similar high throughput studies can be attempted in prokaryotes, though the difficulties surrounding work at the diffraction limit pose challenges, and targeting essential genes in a high throughput way can be difficult. Here we will discuss efforts to make live-cell fluorescent microscopy based experiments using genetically encoded fluorescent reporters an automated, high throughput, and quantitative endeavor amenable to systems-level experiments in bacteria. We emphasize a quantitative data reduction approach, using simulation to help develop biologically relevant cell measurements that completely characterize the cell image. We give an example of how this type of data can be directly exploited by statistical learning algorithms to discover functional pathways.
Figures


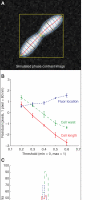
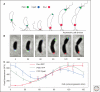
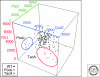

References
-
- Biondi EG, Skerker JM, Arif M, Prasol MS, Perchuk BS, Laub MT 2006. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol Microbiol 59: 386–401 - PubMed
-
- Boland MV, Murphy RF 1999. Automated analysis of patterns in fluorescence-microscope images. Trends Cell Biol 9: 201–202 - PubMed
-
- Boland MV, Murphy RF 2001. A neural network classifier capable of recognizing the patterns of all major subcellular structures in fluorescence microscope images of HeLa cells. Bioinformatics 17: 1213–1223 - PubMed
-
- Boland MV, Markey MK, Murphy RF 1998. Automated recognition of patterns characteristic of subcellular structures in fluorescence microscopy images. Cytometry 33: 366–375 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
