A short N-terminal peptide motif on flavivirus nonstructural protein NS1 modulates cellular targeting and immune recognition
- PMID: 20592095
- PMCID: PMC2937659
- DOI: 10.1128/JVI.00775-10
A short N-terminal peptide motif on flavivirus nonstructural protein NS1 modulates cellular targeting and immune recognition
Abstract
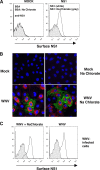
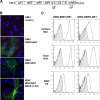
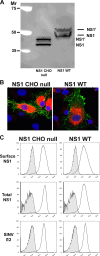
Flavivirus NS1 is a versatile nonstructural glycoprotein, with intracellular NS1 functioning as an essential cofactor for viral replication and cell surface and secreted NS1 antagonizing complement activation. Even though NS1 has multiple functions that contribute to virulence, the genetic determinants that regulate the spatial distribution of NS1 in cells among different flaviviruses remain uncharacterized. Here, by creating a panel of West Nile virus-dengue virus (WNV-DENV) NS1 chimeras and site-specific mutants, we identified a novel, short peptide motif immediately C-terminal to the signal sequence cleavage position that regulates its transit time through the endoplasmic reticulum and differentially directs NS1 for secretion or plasma membrane expression. Exchange of two amino acids within this motif reciprocally changed the cellular targeting pattern of DENV or WNV NS1. For WNV, this substitution also modulated infectivity and antibody-induced phagocytosis of infected cells. Analysis of a mutant lacking all three conserved N-linked glycosylation sites revealed an independent requirement of N-linked glycans for secretion but not for plasma membrane expression of WNV NS1. Collectively, our experiments define the requirements for cellular targeting of NS1, with implications for the protective host responses, immune antagonism, and association with the host cell sorting machinery. These studies also suggest a link between the effects of NS1 on viral replication and the levels of secreted or cell surface NS1.
Figures







Similar articles
-
Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus.J Virol. 2012 Jul;86(13):7360-71. doi: 10.1128/JVI.00157-12. Epub 2012 May 2. J Virol. 2012. PMID: 22553322 Free PMC article.
-
Levels of Circulating NS1 Impact West Nile Virus Spread to the Brain.J Virol. 2021 Sep 27;95(20):e0084421. doi: 10.1128/JVI.00844-21. Epub 2021 Aug 4. J Virol. 2021. PMID: 34346770 Free PMC article.
-
Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation.J Immunol. 2011 Jul 1;187(1):424-33. doi: 10.4049/jimmunol.1100750. Epub 2011 Jun 3. J Immunol. 2011. PMID: 21642539 Free PMC article.
-
Secretion of flaviviral non-structural protein NS1: from diagnosis to pathogenesis.Novartis Found Symp. 2006;277:233-47; discussion 247-53. doi: 10.1002/0470058005.ch17. Novartis Found Symp. 2006. PMID: 17319166 Review.
-
The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker.Antiviral Res. 2013 May;98(2):192-208. doi: 10.1016/j.antiviral.2013.03.008. Epub 2013 Mar 21. Antiviral Res. 2013. PMID: 23523765 Review.
Cited by
-
An alternative -1/+2 open reading frame exists within viral N(pro)(1-19) region of bovine viral diarrhea virus SD-1.Virus Res. 2012 Jan;163(1):341-51. doi: 10.1016/j.virusres.2011.10.022. Epub 2011 Nov 4. Virus Res. 2012. PMID: 22079882 Free PMC article.
-
West Nile Virus NS1 Antagonizes Interferon Beta Production by Targeting RIG-I and MDA5.J Virol. 2017 Aug 24;91(18):e02396-16. doi: 10.1128/JVI.02396-16. Print 2017 Sep 15. J Virol. 2017. PMID: 28659477 Free PMC article.
-
Comparative specificity and sensitivity of NS1-based serological assays for the detection of flavivirus immune response.PLoS Negl Trop Dis. 2020 Jan 29;14(1):e0008039. doi: 10.1371/journal.pntd.0008039. eCollection 2020 Jan. PLoS Negl Trop Dis. 2020. PMID: 31995566 Free PMC article.
-
Hepatitis C Virus core+1/ARF Protein Modulates the Cyclin D1/pRb Pathway and Promotes Carcinogenesis.J Virol. 2018 Apr 13;92(9):e02036-17. doi: 10.1128/JVI.02036-17. Print 2018 May 1. J Virol. 2018. PMID: 29444947 Free PMC article.
-
Genetic characterization of dengue virus serotype 1 circulating in Reunion Island, 2019-2021, and the Seychelles, 2015-2016.BMC Infect Dis. 2023 May 5;23(1):294. doi: 10.1186/s12879-023-08125-y. BMC Infect Dis. 2023. PMID: 37147570 Free PMC article.
References
-
- Alcon-LePoder, S., M. T. Drouet, P. Roux, M. P. Frenkiel, M. Arborio, A. M. Durand-Schneider, M. Maurice, I. Le Blanc, J. Gruenberg, and M. Flamand. 2005. The secreted form of dengue virus nonstructural protein NS1 is endocytosed by hepatocytes and accumulates in late endosomes: implications for viral infectivity. J. Virol. 79:11403-11411. - PMC - PubMed
-
- Alcon-LePoder, S., P. Sivard, M. T. Drouet, A. Talarmin, C. Rice, and M. Flamand. 2006. Secretion of flaviviral non-structural protein NS1: from diagnosis to pathogenesis. Novartis Found. Symp. 277:233-247; discussion 247-253. - PubMed
-
- Avirutnan, P., N. Punyadee, S. Noisakran, C. Komoltri, S. Thiemmeca, K. Auethavornanan, A. Jairungsri, R. Kanlaya, N. Tangthawornchaikul, C. Puttikhunt, S. N. Pattanakitsakul, P. T. Yenchitsomanus, J. Mongkolsapaya, W. Kasinrerk, N. Sittisombut, M. Husmann, M. Blettner, S. Vasanawathana, S. Bhakdi, and P. Malasit. 2006. Vascular leakage in severe dengue virus infections: a potential role for the nonstructural viral protein NS1 and complement. J. Infect. Dis. 193:1078-1088. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

