Evaluation of one- and two-color gene expression arrays for microbial comparative genome hybridization analyses in routine applications
- PMID: 20592156
- PMCID: PMC2937706
- DOI: 10.1128/JCM.00233-10
Evaluation of one- and two-color gene expression arrays for microbial comparative genome hybridization analyses in routine applications
Abstract
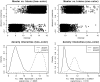
DNA microarray technology has already revolutionized basic research in infectious diseases, and whole-genome sequencing efforts have allowed for the fabrication of tailor-made spotted microarrays for an increasing number of bacterial pathogens. However, the application of microarrays in diagnostic microbiology is currently hampered by the high costs associated with microarray experiments and the specialized equipment needed. Here, we show that a thorough bioinformatic postprocessing of the microarray design to reduce the amount of unspecific noise also allows the reliable use of spotted gene expression microarrays for gene content analyses. We further demonstrate that the use of only single-color labeling to halve the costs for dye-labeled nucleotides results in only a moderate decrease in overall specificity and sensitivity. Therefore, gene expression microarrays using only single-color labeling can also reliably be used for gene content analyses, thus reducing the costs for potential routine applications such as genome-based pathogen detection or strain typing.
Figures

Similar articles
-
Development of a novel ozone- and photo-stable HyPer5 red fluorescent dye for array CGH and microarray gene expression analysis with consistent performance irrespective of environmental conditions.BMC Biotechnol. 2008 Nov 12;8:86. doi: 10.1186/1472-6750-8-86. BMC Biotechnol. 2008. PMID: 19014508 Free PMC article.
-
Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays.BMC Genomics. 2006 Mar 21;7:59. doi: 10.1186/1471-2164-7-59. BMC Genomics. 2006. PMID: 16551369 Free PMC article.
-
RNA and DNA microarrays.Methods Mol Biol. 2011;671:3-34. doi: 10.1007/978-1-59745-551-0_1. Methods Mol Biol. 2011. PMID: 20967621
-
Host-microbe interaction systems biology: lifecycle transcriptomics and comparative genomics.Future Microbiol. 2010 Feb;5(2):205-19. doi: 10.2217/fmb.09.125. Future Microbiol. 2010. PMID: 20143945 Free PMC article. Review.
-
Genomic profiling: cDNA arrays and oligoarrays.Methods Mol Biol. 2012;823:89-105. doi: 10.1007/978-1-60327-216-2_7. Methods Mol Biol. 2012. PMID: 22081341 Review.
Cited by
-
Recent advances in molecular technologies and their application in pathogen detection in foods with particular reference to yersinia.J Pathog. 2011;2011:310135. doi: 10.4061/2011/310135. Epub 2011 Oct 29. J Pathog. 2011. PMID: 22567329 Free PMC article.
-
The zinc-responsive regulon of Neisseria meningitidis comprises 17 genes under control of a Zur element.J Bacteriol. 2012 Dec;194(23):6594-603. doi: 10.1128/JB.01091-12. Epub 2012 Oct 5. J Bacteriol. 2012. PMID: 23043002 Free PMC article.
-
Virulence evolution of the human pathogen Neisseria meningitidis by recombination in the core and accessory genome.PLoS One. 2011 Apr 26;6(4):e18441. doi: 10.1371/journal.pone.0018441. PLoS One. 2011. PMID: 21541312 Free PMC article.
-
Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence.J Bacteriol. 2010 Oct;192(20):5363-77. doi: 10.1128/JB.00883-10. Epub 2010 Aug 13. J Bacteriol. 2010. PMID: 20709895 Free PMC article.
-
Transcriptomic buffering of cryptic genetic variation contributes to meningococcal virulence.BMC Genomics. 2017 Apr 7;18(1):282. doi: 10.1186/s12864-017-3616-7. BMC Genomics. 2017. PMID: 28388876 Free PMC article.
References
-
- Attoor, S., E. R. Dougherty, Y. Chen, M. L. Bittner, and J. M. Trent. 2004. Which is better for cDNA-microarray-based classification: ratios or direct intensities. Bioinformatics 20:2513-2520. - PubMed
-
- Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. - PubMed
-
- Bentley, S. D., G. S. Vernikos, L. A. Snyder, C. Churcher, C. Arrowsmith, T. Chillingworth, A. Cronin, P. H. Davis, N. E. Holroyd, K. Jagels, M. Maddison, S. Moule, E. Rabbinowitsch, S. Sharp, L. Unwin, S. Whitehead, M. A. Quail, M. Achtman, B. Barrell, N. J. Saunders, and J. Parkhill. 2007. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 3:e23. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

