Natural selection and the distribution of identity-by-descent in the human genome
- PMID: 20592267
- PMCID: PMC2940294
- DOI: 10.1534/genetics.110.113977
Natural selection and the distribution of identity-by-descent in the human genome
Abstract
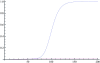
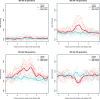
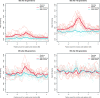
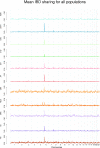
There has recently been considerable interest in detecting natural selection in the human genome. Selection will usually tend to increase identity-by-descent (IBD) among individuals in a population, and many methods for detecting recent and ongoing positive selection indirectly take advantage of this. In this article we show that excess IBD sharing is a general property of natural selection and we show that this fact makes it possible to detect several types of selection including a type that is otherwise difficult to detect: selection acting on standing genetic variation. Motivated by this, we use a recently developed method for identifying IBD sharing among individuals from genome-wide data to scan populations from the new HapMap phase 3 project for regions with excess IBD sharing in order to identify regions in the human genome that have been under strong, very recent selection. The HLA region is by far the region showing the most extreme signal, suggesting that much of the strong recent selection acting on the human genome has been immune related and acting on HLA loci. As equilibrium overdominance does not tend to increase IBD, we argue that this type of selection cannot explain our observations.
Figures



References
-
- Albrechtsen, A., T. Sand Korneliussen, I. Moltke, T. van Overseem Hansen, F. C. Nielsen et al., 2009. Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium. Genet. Epidemiol. 33 266–274. - PubMed
-
- Barrett, R. D., and D. Schluter, 2008. Adaptation from standing genetic variation. Trends Ecol. Evol. 23 38–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

