Development of an immunocapture-polymerase chain reaction assay using IgY to detect Mycobacterium avium subsp. paratuberculosis
- PMID: 20592839
- PMCID: PMC2851719
Development of an immunocapture-polymerase chain reaction assay using IgY to detect Mycobacterium avium subsp. paratuberculosis
Abstract
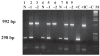
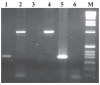
A diagnostic assay using immunomagnetic separation was developed to capture Mycobacterium avium subsp. paratuberculosis (MAP) from bovine feces by means of IgY derived from chicken eggs. The antibody was coupled directly onto the surface of MagaCell cellulose/iron oxide beads or indirectly by being mixed with MagaBeads and a rabbit IgG linker against chicken antigen. Optimization parameters for the immunocapture included incubation time, temperature, volume, and type of immunocapture beads. Analytical sensitivity and specificity were determined by extracting DNA from the captured bacteria and amplifying it by polymerase chain reaction (PCR). The 2 bead preparations had the same analytical sensitivity, and the detection level of MAP cells in spiked bovine feces was 2 x 10(4) cells/g. No PCR inhibition was observed with DNA from the organisms captured with use of the MagaCell-IgY beads.
Une épreuve diagnostique utilisant la séparation immuno-magnétique a été développée afin de capturer Mycobacterium avium ssp. paratuberculosis (MAP) à partir de fèces bovines au moyen d’IgY provenant d’œufs de poule. Les anticorps ont été couplés directement sur la surface de billes MagaCell composées de cellullose/oxyde de fer ou indirectement en étant mélangées avec des MagaBeads et des IgG de lapin dirigés contre les antigènes de poulet. L’optimisation des paramètres pour l’immunocapture incluait le temps d’incubation, la température, le volume et le type de billes. La sensibilité analytique et la spécificité ont été déterminées par extraction de l’ADN des bactéries capturées et en l’amplifiant par réaction d’amplification en chaîne par la polymérase (PCR). Les 2 préparations de billes avaient la même sensibilité analytique, et le degré de détection des cellules de MAP dans des échantillons inoculés intentionnellement était de 2 × 104 cellules/g de fèces bovines. Aucune inhibition du PCR n’a été observée avec l’ADN des microorganismes capturés en utilisant les billes MagaCell-IgY.
(Traduit par Docteur Serge Messier)
Figures



Similar articles
-
Rapid and sensitive detection of Mycobacterium avium subsp. paratuberculosis in bovine milk and feces by a combination of immunomagnetic bead separation-conventional PCR and real-time PCR.J Clin Microbiol. 2004 Mar;42(3):1075-81. doi: 10.1128/JCM.42.3.1075-1081.2004. J Clin Microbiol. 2004. PMID: 15004056 Free PMC article.
-
Comparison of immunomagnetic bead separation-polymerase chain reaction and faecal culture for the detection of Mycobacterium avium subsp paratuberculosis in sheep faeces.Aust Vet J. 2001 Jul;79(7):497-500. doi: 10.1111/j.1751-0813.2001.tb13024.x. Aust Vet J. 2001. PMID: 11549050 No abstract available.
-
Improved detection of Mycobacterium avium subsp. paratuberculosis In milk by immunomagnetic PCR.Vet Microbiol. 2000 Dec 20;77(3-4):369-78. doi: 10.1016/s0378-1135(00)00322-9. Vet Microbiol. 2000. PMID: 11118722
-
Comparison of Immunomagnetic Bead Separation-Immunosensor Detection and Nested-PCR Methods for Detecting Mycobacterium avium Subspecies paratuberculosis in Cattle Feces.J Clin Lab Anal. 2025 Apr;39(7):e70009. doi: 10.1002/jcla.70009. Epub 2025 Mar 19. J Clin Lab Anal. 2025. PMID: 40105275 Free PMC article.
-
New triplex real-time PCR assay for detection of Mycobacterium avium subsp. paratuberculosis in bovine feces.Appl Environ Microbiol. 2008 May;74(9):2751-8. doi: 10.1128/AEM.02534-07. Epub 2008 Mar 7. Appl Environ Microbiol. 2008. PMID: 18326682 Free PMC article.
Cited by
-
Management of Mycobacterium avium subsp. paratuberculosis in dairy farms: Selection and evaluation of different DNA extraction methods from bovine and buffaloes milk and colostrum for the establishment of a safe colostrum farm bank.Microbiologyopen. 2019 Oct;8(10):e875. doi: 10.1002/mbo3.875. Epub 2019 Aug 17. Microbiologyopen. 2019. PMID: 31420952 Free PMC article.
-
Mycobacterium paratuberculosis detection in cow's milk in Argentina by immunomagnetic separation-PCR.Braz J Microbiol. 2016 Apr-Jun;47(2):506-12. doi: 10.1016/j.bjm.2016.01.013. Epub 2016 Mar 2. Braz J Microbiol. 2016. PMID: 26991290 Free PMC article.
-
Detection of Mycobacterium avium Subspecies paratuberculosis (MAP) Microorganisms Using Antigenic MAP Cell Envelope Proteins.Front Vet Sci. 2021 Feb 3;8:615029. doi: 10.3389/fvets.2021.615029. eCollection 2021. Front Vet Sci. 2021. PMID: 33614761 Free PMC article.
References
-
- Chiodini RJ, Van Kruiningen HJ, Merkal RS. Ruminant paratuberculosis (Johne’s disease): The current status and future prospects. Cornell Vet. 1984;74:218–262. - PubMed
-
- Riemann H, Zaman MR, Ruppanner R, et al. Paratuberculosis in cattle and free-living exotic deer. J Am Vet Med Assoc. 1979;174:841–843. - PubMed
-
- Beard PM, Daniels MJ, Henderson D. Evidence of paratuberculosis in fox (Vulpes vuples) and stoat (Mustela erminea) Vet Rec. 1999;145:612–613.
-
- Greig A, Stevenson K, Perez V, Pirie AA, Grant JM, Sharp JM. Paratuberculosis in wild rabbits (Oryctolagus cuniculus) Vet Rec. 1997;140:141–143. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
