A major histocompatibility Class I locus contributes to multiple sclerosis susceptibility independently from HLA-DRB1*15:01
- PMID: 20593013
- PMCID: PMC2892470
- DOI: 10.1371/journal.pone.0011296
A major histocompatibility Class I locus contributes to multiple sclerosis susceptibility independently from HLA-DRB1*15:01
Abstract
Background: In Northern European descended populations, genetic susceptibility for multiple sclerosis (MS) is associated with alleles of the human leukocyte antigen (HLA) Class II gene DRB1. Whether other major histocompatibility complex (MHC) genes contribute to MS susceptibility is controversial.
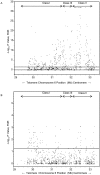
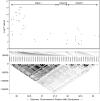
Methodology/principal findings: A case control analysis was performed using 958 single nucleotide polymorphisms (SNPs) spanning the MHC assayed in two independent datasets. The discovery dataset consisted of 1,018 cases and 1,795 controls and the replication dataset was composed of 1,343 cases and 1,379 controls. The most significantly MS-associated SNP in the discovery dataset was rs3135391, a Class II SNP known to tag the HLA-DRB1*15:01 allele, the primary MS susceptibility allele in the MHC (O.R. = 3.04, p < 1 x 10(-78)). To control for the effects of the HLA-DRB1*15:01 haplotype, case control analysis was performed adjusting for this HLA-DRB1*15:01 tagging SNP. After correction for multiple comparisons (false discovery rate = .05) 52 SNPs in the Class I, II and III regions were significantly associated with MS susceptibility in both datasets using the Cochran Armitage trend test. The discovery and replication datasets were merged and subjects carrying the HLA-DRB1*15:01 tagging SNP were excluded. Association tests showed that 48 of the 52 replicated SNPs retained significant associations with MS susceptibility independently of the HLA-DRB1*15:01 as defined by the tagging SNP. 20 Class I SNPs were associated with MS susceptibility with p-values < or = 1 x 10(-8). The most significantly associated SNP was rs4959039, a SNP in the downstream un-translated region of the non-classical HLA-G gene (Odds ratio 1.59, 95% CI 1.40, 1.81, p = 8.45 x 10(-13)) and is in linkage disequilibrium with several nearby SNPs. Logistic regression modeling showed that this SNP's contribution to MS susceptibility was independent of the Class II and Class III SNPs identified in this screen.
Conclusions: A MHC Class I locus contributes to MS susceptibility independently of the HLA-DRB1*15:01 haplotype.
Conflict of interest statement
Figures
References
-
- Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. - PubMed
-
- Barcellos LF, Oksenberg JR, Green AJ, Bucher P, Rimmler JB, et al. Genetic basis for clinical expression in multiple sclerosis. Brain. 2002;125:150–158. - PubMed
-
- Lincoln MR, Montpetit A, Cader MZ, Saarela J, Dyment DA, et al. A predominant role for the HLA class II region in the association of the MHC region with multiple sclerosis. Nat Genet. 2005;37:1108–1112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

