Antagonistic changes in sensitivity to antifungal drugs by mutations of an important ABC transporter gene in a fungal pathogen
- PMID: 20593017
- PMCID: PMC2892482
- DOI: 10.1371/journal.pone.0011309
Antagonistic changes in sensitivity to antifungal drugs by mutations of an important ABC transporter gene in a fungal pathogen
Abstract
Fungal pathogens can be lethal, especially among immunocompromised populations, such as patients with AIDS and recipients of tissue transplantation or chemotherapy. Prolonged usage of antifungal reagents can lead to drug resistance and treatment failure. Understanding mechanisms that underlie drug resistance by pathogenic microorganisms is thus vital for dealing with this emerging issue. In this study, we show that dramatic sequence changes in PDR5, an ABC (ATP-binding cassette) efflux transporter protein gene in an opportunistic fungal pathogen, caused the organism to become hypersensitive to azole, a widely used antifungal drug. Surprisingly, the same mutations conferred growth advantages to the organism on polyenes, which are also commonly used antimycotics. Our results indicate that Pdr5p might be important for ergosterol homeostasis. The observed remarkable sequence divergence in the PDR5 gene in yeast strain YJM789 may represent an interesting case of adaptive loss of gene function with significant clinical implications.
Conflict of interest statement
Figures
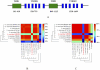

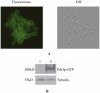



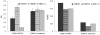
Similar articles
-
β-Lapachone enhances the antifungal activity of fluconazole against a Pdr5p-mediated resistant Saccharomyces cerevisiae strain.Braz J Microbiol. 2020 Sep;51(3):1051-1060. doi: 10.1007/s42770-020-00254-9. Epub 2020 Mar 10. Braz J Microbiol. 2020. PMID: 32157667 Free PMC article.
-
Characterization of the Efflux Capability and Substrate Specificity of Aspergillus fumigatus PDR5-like ABC Transporters Expressed in Saccharomyces cerevisiae.mBio. 2020 Mar 24;11(2):e00338-20. doi: 10.1128/mBio.00338-20. mBio. 2020. PMID: 32209680 Free PMC article.
-
Functional analysis of an ATP-binding cassette transporter protein from Aspergillus fumigatus by heterologous expression in Saccharomyces cerevisiae.Fungal Genet Biol. 2013 Aug;57:85-91. doi: 10.1016/j.fgb.2013.06.004. Epub 2013 Jun 21. Fungal Genet Biol. 2013. PMID: 23796749 Free PMC article.
-
Membrane transporters and antifungal drug resistance.Curr Drug Targets. 2000 Nov;1(3):261-84. doi: 10.2174/1389450003349209. Curr Drug Targets. 2000. PMID: 11465075 Review.
-
Characterization of the multi-drug efflux systems of pathogenic fungi using functional hyperexpression in Saccharomyces cerevisiae.Nihon Ishinkin Gakkai Zasshi. 2010;51(2):79-86. doi: 10.3314/jjmm.51.79. Nihon Ishinkin Gakkai Zasshi. 2010. PMID: 20467195 Review.
Cited by
-
Resistance Mechanisms of Saccharomyces cerevisiae to Commercial Formulations of Glyphosate Involve DNA Damage Repair, the Cell Cycle, and the Cell Wall Structure.G3 (Bethesda). 2020 Jun 1;10(6):2043-2056. doi: 10.1534/g3.120.401183. G3 (Bethesda). 2020. PMID: 32299824 Free PMC article.
-
Divergence in a master variator generates distinct phenotypes and transcriptional responses.Genes Dev. 2014 Feb 15;28(4):409-21. doi: 10.1101/gad.228940.113. Genes Dev. 2014. PMID: 24532717 Free PMC article.
-
ABC Transporter Genes Show Upregulated Expression in Drug-Resistant Clinical Isolates of Candida auris: A Genome-Wide Characterization of ATP-Binding Cassette (ABC) Transporter Genes.Front Microbiol. 2019 Jul 16;10:1445. doi: 10.3389/fmicb.2019.01445. eCollection 2019. Front Microbiol. 2019. PMID: 31379756 Free PMC article.
-
Yeast of Eden: microbial resistance to glyphosate from a yeast perspective.Curr Genet. 2023 Dec;69(4-6):203-212. doi: 10.1007/s00294-023-01272-4. Epub 2023 Jun 3. Curr Genet. 2023. PMID: 37269314 Free PMC article. Review.
-
Control of Plasma Membrane Permeability by ABC Transporters.Eukaryot Cell. 2015 May;14(5):442-53. doi: 10.1128/EC.00021-15. Epub 2015 Feb 27. Eukaryot Cell. 2015. PMID: 25724885 Free PMC article.
References
-
- Groll AH, Tragiannidis A. Recent advances in antifungal prevention and treatment. Semin Hematol. 2009;46:212–229. - PubMed
-
- Sanglard D, Coste A, Ferrari S. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 2009;9:1029–1050. - PubMed
-
- Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. - PubMed
-
- Goffeau A. Drug resistance: The fight against fungi. Nature. 2008;452:541–542. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

