Protein-protein interactions between sigma(70) region 4 of RNA polymerase and Escherichia coli SoxS, a transcription activator that functions by the prerecruitment mechanism: evidence for "off-DNA" and "on-DNA" interactions
- PMID: 20595001
- PMCID: PMC2917807
- DOI: 10.1016/j.jmb.2010.05.052
Protein-protein interactions between sigma(70) region 4 of RNA polymerase and Escherichia coli SoxS, a transcription activator that functions by the prerecruitment mechanism: evidence for "off-DNA" and "on-DNA" interactions
Abstract
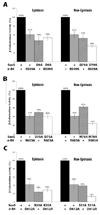
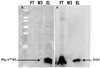
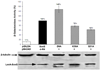
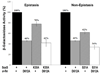
According to the prerecruitment hypothesis, Escherichia coli SoxS activates the transcription of the genes of the SoxRS regulon by forming binary complexes with RNA polymerase (RNAP) that scan the chromosome for class I and class II SoxS-dependent promoters. We showed previously that the alpha subunit's C-terminal domain plays a role in activating both classes of promoter by making protein-protein contacts with SoxS; some of these contacts are made in solution in the absence of promoter DNA, a critical prediction of the prerecruitment hypothesis. Here, we identified seven single-alanine substitutions of the region 4 of sigma(70) (sigma(70) R4) of RNAP that reduce SoxS activation of class II promoters. With genetic epistasis tests between these sigma(70) R4 mutants and positive control mutants of SoxS, we identified 10 pairs of amino acids that interact with each other in E. coli. Using the yeast two-hybrid system and affinity immobilization assays, we showed that SoxS and sigma(70) R4 can interact in solution (i.e., "off-DNA"). The interaction requires amino acids of the class I/II (but not the class II) positive control surface of SoxS, and five amino acids of sigma(70) R4 that reduce activation in E. coli also reduce the SoxS-sigma(70) R4 interaction in yeast. One of the epistatic interactions that occur in E. coli also occurs in the yeast two-hybrid system (i.e., off-DNA). Importantly, we infer that the five epistatic interactions occurring in E. coli that require an amino acid of the class II surface occur "on-DNA" at class II promoters. Finding that SoxS contacts sigma(70) R4 both off-DNA and on-DNA is consistent with the prerecruitment hypothesis. Moreover, SoxS is now the first example of an E. coli transcriptional activator that uses a single positive control surface to make specific protein-protein contacts with two different subunits of RNAP.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Transcription activation by Escherichia coli Rob at class II promoters: protein-protein interactions between Rob's N-terminal domain and the σ(70) subunit of RNA polymerase.J Mol Biol. 2012 Jun 8;419(3-4):139-57. doi: 10.1016/j.jmb.2012.03.019. Epub 2012 Mar 28. J Mol Biol. 2012. PMID: 22465792 Free PMC article.
-
Genetic evidence for a novel interaction between transcriptional activator SoxS and region 4 of the σ(70) subunit of RNA polymerase at class II SoxS-dependent promoters in Escherichia coli.J Mol Biol. 2011 Apr 1;407(3):333-53. doi: 10.1016/j.jmb.2010.12.037. Epub 2010 Dec 31. J Mol Biol. 2011. PMID: 21195716 Free PMC article.
-
Novel protein--protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA polymerase alpha subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promoters during oxidative stress.J Mol Biol. 2004 Oct 22;343(3):513-32. doi: 10.1016/j.jmb.2004.08.057. J Mol Biol. 2004. PMID: 15465042
-
Structure and function of RapA: a bacterial Swi2/Snf2 protein required for RNA polymerase recycling in transcription.Biochim Biophys Acta. 2011 Sep;1809(9):470-5. doi: 10.1016/j.bbagrm.2011.03.003. Epub 2011 Mar 16. Biochim Biophys Acta. 2011. PMID: 21419241 Free PMC article. Review.
-
Mechanisms for activating bacterial RNA polymerase.FEMS Microbiol Rev. 2010 Sep;34(5):611-27. doi: 10.1111/j.1574-6976.2010.00239.x. Epub 2010 Jun 7. FEMS Microbiol Rev. 2010. PMID: 20629756 Review.
Cited by
-
Coordination of cell envelope biology by Escherichia coli MarA protein potentiates intrinsic antibiotic resistance.PLoS Genet. 2025 May 5;21(5):e1011639. doi: 10.1371/journal.pgen.1011639. eCollection 2025 May. PLoS Genet. 2025. PMID: 40324004 Free PMC article.
-
Transcription activation by Escherichia coli Rob at class II promoters: protein-protein interactions between Rob's N-terminal domain and the σ(70) subunit of RNA polymerase.J Mol Biol. 2012 Jun 8;419(3-4):139-57. doi: 10.1016/j.jmb.2012.03.019. Epub 2012 Mar 28. J Mol Biol. 2012. PMID: 22465792 Free PMC article.
-
Structural basis of three different transcription activation strategies adopted by a single regulator SoxS.Nucleic Acids Res. 2022 Oct 28;50(19):11359-11373. doi: 10.1093/nar/gkac898. Nucleic Acids Res. 2022. PMID: 36243985 Free PMC article.
-
Structural basis of non-canonical transcriptional regulation by the σA-bound iron-sulfur protein WhiB1 in M. tuberculosis.Nucleic Acids Res. 2020 Jan 24;48(2):501-516. doi: 10.1093/nar/gkz1133. Nucleic Acids Res. 2020. PMID: 31807774 Free PMC article.
-
Repurposing endogenous type I-E CRISPR-Cas systems for natural product discovery in Streptomyces.Nat Commun. 2024 Nov 13;15(1):9833. doi: 10.1038/s41467-024-54196-z. Nat Commun. 2024. PMID: 39537651 Free PMC article.
References
-
- Burgess RR. Separation and characterization of the subunits of RNA polymerase. J. Biol. Chem. 1969;244:6168–6178. - PubMed
-
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. - PubMed
-
- Burgess RR, Travers AA, Dunn JJ, Bautz EKF. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. - PubMed
-
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. - PubMed
-
- Korzheva N, Mustaev A. Transcription elongation complex: structure and function. Curr. Opin. Microbiol. 2001;4:119–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

